Page 45 - EJMO-9-3
P. 45
Eurasian Journal of
Medicine and Oncology NOTCH3 in CSVD and breast cancer
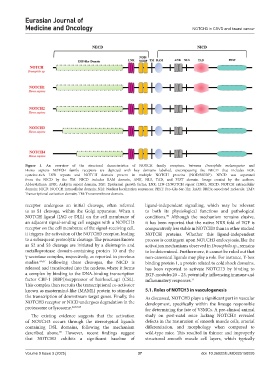
Figure 1. An overview of the structural characteristics of NOTCH family receptors, between Drosophila melanogaster and
Homo sapiens NOTCH family receptors are depicted with key domains labelled, encompassing the NECD that includes EGF,
cysteine-rich LNR repeats and NOTCH domain present in multiple NOTCH proteins (NOD/NODP). NECD was separated
from the NICD by the TM. NICD includes RAM domain, ANK, NLS, TAD, and PEST domain. Image created by the authors.
Abbreviations: ANK: Ankyrin repeat domain; EGF: Epidermal growth factor; LNR: LIN-12/NOTCH repeat (LNR); NECD: NOTCH extracellular
domain; NICD: NOTCH intracellular domain; NLS: Nuclear localization sequences; PEST: Pro-Glu-Ser-Thr; RAM: RBPJκ-associated molecule; TAD:
Transcriptional activation domain; TM: Transmembrane domain.
receptor undergoes an initial cleavage, often referred ligand-independent signalling, which may be relevant
to as S1 cleavage, within the Golgi apparatus. When a to both its physiological functions and pathological
NOTCH ligand (JAG or DLL) on the cell membrane of conditions. Although the mechanism remains elusive,
70
an adjacent signal-sending cell engages with a NOTCH3 it has been reported that the native NRR fold of EGF is
receptor on the cell membrane of the signal-receiving cell, comparatively less stable in NOTCH3 than in other studied
it triggers the activation of the NOTCH3 receptor, leading NOTCH proteins. Whether this ligand-independent
to a subsequent proteolytic cleavage. The processes known process is contingent upon NOTCH3 endocytosis, like the
as S2 and S3 cleavage are initiated by a disintegrin and activation mechanisms observed in Drosophila sp., remains
metalloprotease domain-containing protein 10 and the to be determined. Furthermore, it cannot be ruled out that
γ-secretase complex, respectively, as reported in previous non-canonical ligands may play a role. For instance, Y-box
studies. 62,67 Following these cleavages, the NICD is binding protein 1, a protein related to cold shock domains,
released and translocated into the nucleus, where it forms has been reported to activate NOTCH3 by binding to
a complex by binding to the DNA-binding transcription EGF-modules 20 – 23, potentially influencing immune and
factor CBF-1 (RBPJ)/suppressor of hairless/Lag1 (CSL). inflammatory responses. 71
This complex then recruits the transcriptional co-activator
known as mastermind-like (MAML) protein to stimulate 5.1. Roles of NOTCH3 in vasculogenesis
the transcription of downstream target genes. Finally, the As discussed, NOTCH3 plays a significant part in vascular
NOTCH3 receptor or NICD undergoes degradation in the development, specifically within the lineage responsible
proteasome or lysosome. 62,67,68 for determining the fate of VSMCs. A pre-clinical animal
The existing evidence suggests that the activation study on post-natal mice lacking NOTCH3 revealed
of NOTCH3 occurs through the stereotypical ligands defects in the maturation of smooth muscle cells, arterial
containing DSL domains, following the mechanism differentiation, and morphology when compared to
described above. However, recent findings suggest wild-type mice. This resulted in thinner and improperly
69
that NOTCH3 exhibits a significant baseline of structured smooth muscle cell layers, which typically
Volume 9 Issue 3 (2025) 37 doi: 10.36922/EJMO025150095