Page 39 - GPD-4-2
P. 39
Gene & Protein in Disease Orexin in depression
antidepressant therapies. The cAMP-response element- by orexin control energy metabolism and sleep and are
binding protein (CREB) is a transcription factor binding associated with depression.
to specific DNA sequences and regulating gene expression. Orexin neurons critically modulate arousal and reward
The CREB signaling pathway has been identified as a key processing by interacting with key neurotransmitter
intracellular mechanism influenced by orexin, which systems, remarkably the dopamine and GABAergic
may clarify its impact on energy homeostasis and the pathways. Specifically, the influence of orexin on the
13
sleep–wake cycle. Orexin can affect the CREB activity by CREB pathway emphasizes its potential to regulate energy
activating intracellular signaling pathways. In an inactive balance and arousal at the cellular level. The activation of
state, CREB may be present in the cytoplasm; however, orexin receptors modulates synaptic plasticity, influencing
on stimulation by specific signals, such as activation by mood-related behaviors through direct interactions
the cAMP/PKA (protein kinase A) pathway, CREB is with the hypothalamic-pituitary-adrenal (HPA) axis
10
phosphorylated and translocated to the nucleus, where it (Figure 5) and other critical stress-response circuits; 14,15
plays a remarkable role in regulating energy homeostasis this complex interplay may underlie the observed decrease
and wakefulness (Figure 3). This indicates that orexin in orexin levels in depressive states, thereby linking orexin
is essential for maintaining a state of arousal. Orexin dysregulation to depression pathophysiology.
11
neurons regulate various physiological activities through This review article aims to provide novel insights
interactions with monoaminergic, dynorphin, and into the use of orexin in the treating and preventing
serotonergic neurons (Figure 4). Transcripts controlled depression. This article discusses orexin expression levels,
12
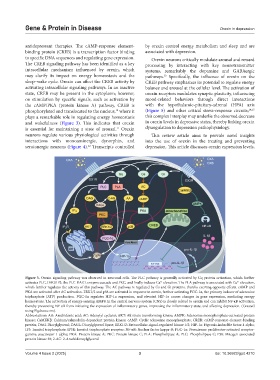
Figure 3. Orexin signaling pathway was observed in neuronal cells. The PLC pathway is generally activated by Gq protein activation, which further
activates PLC, DAG/IP3, the PLC-DAGL enzyme cascade and PKC, and finally induces Ca elevation. The PLA pathway is associated with Ca elevation,
2+
2+
which further regulates the activity of this pathway. The AC pathway is regulated by Gs and Gi proteins, thereby exerting opposite effects. cAMP and
PKA are activated after AC activation. ERK1/2 and p38 are activated in response to orexin, further activating PGC-1α, the primary inducer of adenosine
triphosphate (ATP) production. PGC-1α regulates HIF-1α expression, and elevated HIF-1α causes changes in gene expression, mediating energy
homeostasis. The activation of energy-sensing AMPK in the central nervous system (CNS) is closely related to orexin and can inhibit NF-κB activation,
thereby preventing NF-κB from initiating the expression of inflammatory genes, improving the inflammatory state, and affecting depression. (Created
using Figdraw.com).
Abbreviations: AA: Arachidonic acid; AC: Adenylyl cyclases; AKT: Ak strain transforming kinase; AMPK: Adenosine monophosphate-activated protein
kinase; CaMKKβ: Calcium/calmodulin-dependent protein kinase; cAMP: Cyclic adenosine monophosphate; CREB: cAMP-response element-binding
protein; DAG: Diacylglycerol; DAGL: Diacylglycerol lipase; ERK1/2: Extracellular-signal-regulated kinase 1/2; HIF-1α: Hypoxia-inducible factor-1 alpha;
IP3: Inositol trisphosphate; IP3R: Inositol trisphosphate receptor; NF-κB: Nuclear factor kappa-B; PGC-1α: Peroxisome proliferator-activated receptor-
gamma coactivator 1-alpha; PKA: Protein kinase A; PKC: Protein kinase C; PLA: Phospholipase A; PLC: Phospholipase C; P38: Mitogen-associated
protein kinase 38; 2-AG: 2-Arachidonoylglycerol.
Volume 4 Issue 2 (2025) 3 doi: 10.36922/gpd.4210