Page 108 - MI-2-4
P. 108
Microbes & Immunity Brachyspira pilosicoli novel outer membrane proteins
A B C D E
F G H I
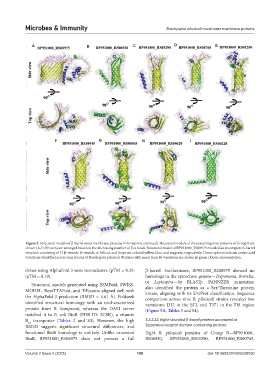
Figure 5. Structural models of β-barrel outer membrane proteins in Group B (continued). Structural models of the remaining nine proteins of Group B are
shown (A-I). Proteins are arranged based on the decreasing number of β-strands. Structural model of BP951000_RS08975 revealed an incomplete β-barrel
structure consisting of 13 β-strands. β-strands, α-helices, and loops are colored yellow, blue, and magenta, respectively. Green spheres indicate amino acid
variations identified across nine strains of Brachyspira pilosicoli. Proteins with more than 40 variations are shown in green ribbon representation.
dimer using AlphaFold 3 were inconclusive (pTM = 0.35; β-barrel. Furthermore, BP951000_RS08975 showed no
ipTM = 0.19). homologs in the spirochete genera—Treponema, Borrelia,
or Leptospira—by BLASTp. PANNZER annotation
Structural models generated using ESMFold, SWISS-
MODEL, RoseTTAFold, and TrRosetta aligned well with also identified the protein as a Ser/Threonine protein
kinase, aligning with its UniProt classification. Sequence
the AlphaFold 3 prediction (RMSD = 4.61 Å). Foldseek comparison across nine B. pilosicoli strains revealed two
identified structural homology with an uncharacterized variations: D32 in the ECL and T371 in the TM region
protein from B. hampsonii, whereas the DALI server (Figure 5A, Tables 3 and S4).
matched it to E. coli BtuB (PDB ID: 2GSK), a vitamin
B transporter (Tables 2 and S3). However, the high 3.2.2.22. Eight-stranded β-barrel proteins annotated as
12
RMSD suggests significant structural differences, and Serpentine receptor domain-containing proteins
functional BtuB homology is unlikely. Unlike canonical Eight B. pilosicoli proteins of Group B—BP951000_
BtuB, BP951000_RS08975 does not possess a full RS06930, BP951000_RS03290, BP951000_RS00765,
Volume 2 Issue 4 (2025) 100 doi: 10.36922/MI025230050