Page 71 - MI-2-4
P. 71
Microbes & Immunity Bioinformatics analysis platform
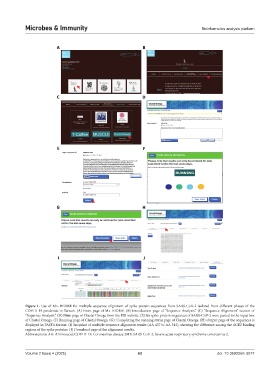
Figure 1. Use of Mx. BIOME for multiple sequence alignment of spike protein sequences from SARS-CoV-2 isolated from different phases of the
COVID-19 pandemic in Taiwan. (A) Front page of Mx. BIOME. (B) Introduction page of “Sequence Analysis.” (C) “Sequence Alignment” section of
“Sequence Analysis.” (D) Main page of Clustal Omega from the EBI website. (E) Six spike protein sequences of SARS-CoV-2 were pasted to the input box
of Clustal Omega. (F) Running page of Clustal Omega. (G): Completing the running status page of Clustal Omega. (H) Output page of the sequences is
displayed in FASTA format. (I) Snapshot of multiple sequence alignment results (AA 437 to AA 512), showing the difference among the ACE2 binding
regions of the spike proteins. (J) Download page of the alignment results.
Abbreviations: AA: Amino acid; COVID-19: Coronavirus disease 2019; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.
Volume 2 Issue 4 (2025) 63 doi: 10.36922/mi.5077