Page 95 - AIH-1-4
P. 95
Artificial Intelligence in Health Transformer-based radiology report summaries
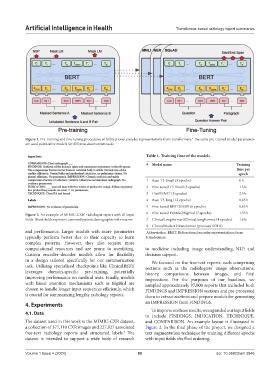
Figure 1. Pre-training and fine-tuning procedures of bidirectional encoder representations from transformers. The same pre-trained model parameters
6
are used to initialize models for different downstream tasks
Table 1. Training time of the models
# Model name Training
time per
epoch
1 Base-T5-Small (3 epochs) 0 h
2 Fine-tuned T5-Small (3 epochs) 1.5 h
3 DistillBART (3 epochs) 2.5 h
4 Base-T5-long (12 epochs) 0.63 h
5 Fine-tuned BERT2BERT (6 epochs) 0.65 h
Figure 2. An example of MIMIC-CXR radiologist report with all input 6 Fine-tuned PubMed BigBird (7 epochs) 1.55 h
3
fields. Blank fields represent censored patient demographic information 7 ClinicalLongFormer2ClinicalLongFormer (4 epochs) 1.6 h
8 ClinicalBioBert2Transformer (previous SOTA) -
and performance. Larger models with more parameters Abbreviation: BERT: Bidirectional encoder representations from
typically perform better due to their capacity to learn transformers.
complex patterns. However, they also require more
computational resources and are prone to overfitting. in medicine including image understanding, NLP, and
Custom encoder–decoder models allow for flexibility decision support.
in a design tailored specifically for our summarization We focused on the free-text reports, each comprising
task. Utilizing specialized checkpoints like ClinicalBERT sections such as the radiologists’ image observations,
leverages domain-specific pre-training, potentially history, comparisons between images, and final
improving performance on medical texts. Finally, models impressions. For the purposes of our baselines, we
with linear attention mechanisms such as BigBird are sampled approximately 97,000 reports that included both
chosen to handle longer input sequences efficiently, which FINDINGS and IMPRESSION sections and pre-processed
is crucial for summarizing lengthy radiology reports. them to extract sections and prepare models for generating
4. Experiments an IMPRESSION from FINDINGS.
To improve on these results, we expanded our input fields
4.1. Data
to include FINDINGS, INDICATION, TECHNIQUE,
The dataset used in this work is the MIMIC-CXR dataset, and COMPARISON. An example layout is illustrated in
a collection of 377,110 CXR images and 227,827 associated Figure 2. In the final phase of the project, we designed a
free-text radiology reports and structured labels. The text augmentation technique by training different epochs
3
dataset is intended to support a wide body of research with input fields shuffled ordering.
Volume 1 Issue 4 (2024) 89 doi: 10.36922/aih.3846