Page 41 - AJWEP-v22i2
P. 41
Cadmium binding and soil microbial diversity in Vetiveria zizanioides
libraries were prepared using the TruSeq DNA
PCR-Free Sample Prep Kit (Illumina, USA) following
the manufacturer’s guidelines. The quality of the library
was assessed using the Qubit 2.0 Fluorometer (Thermo
Scientific) and the Agilent Bioanalyzer 2100 system.
Finally, the library was sequenced on the Illumina
HiSeq 2500 platform, generating 250 bp paired-end
reads (Beijing Genomics Institute, China).
2.3.3. Statistical and bioinformatics analysis
In addition, the sequences were aligned and compared
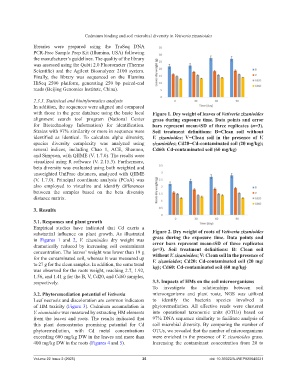
with those in the gene database using the basic local Figure 1. Dry weight of leaves of Vetiveria zizanioides
alignment search tool program (National Center grass during exposure time. Data points and error
for Biotechnology Information) for identification. bars represent mean±SD of three replicates (n=3).
Strains with 97% similarity or more in sequence were Soil treatment definitions: B=Clean soil without
identified as identical. To calculate alpha diversity, V. zizanioides; V=Clean soil in the presence of V.
species diversity complexity was analyzed using zizanioides; Cd20=Cd-contaminated soil (20 mg/kg);
several indices, including Chao 1, ACE, Shannon, Cd60: Cd-contaminated soil (60 mg/kg)
and Simpson, with QIIME (V. 1.7.0). The results were
visualized using R software (V. 2.15.3). Furthermore,
beta diversity was evaluated using both weighted and
unweighted UniFrac distances, analyzed with QIIME
(V. 1.7.0). Principal coordinate analysis (PCoA) was
also employed to visualize and identify differences
between the samples based on the beta diversity
distance matrix.
3. Results
3.1. Responses and plant growth
Empirical studies have indicated that Cd exerts a
substantial influence on plant growth. As illustrated Figure 2. Dry weight of roots of Vetiveria zizanioides
in Figures 1 and 2, V. zizanioides dry weight was grass during the exposure time. Data points and
dramatically reduced by increasing soil contaminant error bars represent mean±SD of three replicates
concentration. The leaves’ weight was lower than 19 g (n=3). Soil treatment definitions: B: Clean soil
without V. zizanioides; V: Clean soil in the presence of
for the contaminated soil, whereas it was measured up V. zizanioides; Cd20: Cd-contaminated soil (20 mg/
to 27 g for the clean samples. In addition, the same trend kg); Cd60: Cd-contaminated soil (60 mg/kg)
was observed for the roots weight, reaching 2.7, 1.92,
1.56, and 1.41 g for the B, V, Cd20, and Cd60 samples,
respectively. 3.3. Impacts of HMs on the soil microorganisms
To investigate the relationships between soil
3.2. Phytoremediation potential of Vetiveria microorganisms and plant roots, NGS was utilized
Leaf necrosis and discoloration are common indicators to identify the bacteria species involved in
of HM toxicity (Figure 3). Cadmium accumulation in phytoremediation. All effective reads were clustered
V. zizanioides was measured by extracting HM elements into operational taxonomic units (OTUs) based on
from the leaves and roots. The results indicated that 97% DNA sequence similarity to facilitate analysis of
this plant demonstrates promising potential for Cd soil microbial diversity. By comparing the number of
phytoremediation, with Cd metal concentrations OTUs, we revealed that the number of microorganisms
exceeding 600 mg/kg DW in the leaves and more than were enriched in the presence of V. zizanioides grass.
400 mg/kg DW in the roots (Figures 4 and 5). Increasing the contaminant concentration from 20 to
Volume 22 Issue 2 (2025) 35 doi: 10.36922/AJWEP025040021