Page 154 - EJMO-9-1
P. 154
Eurasian Journal of Medicine and
Oncology
Potential of flavonoids against glioblastoma
2. Methods
2.1. Plant collection
The bark of P. chinensis was collected from the area
surrounding Hostel 02 at the University of Peshawar,
Khyber Pakhtunkhwa, Pakistan. To ensure the authenticity
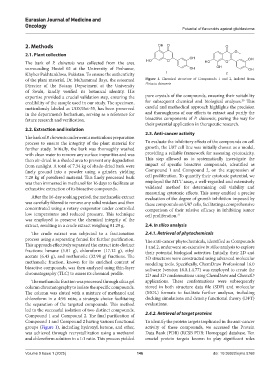
of the plant material, Dr. Muhammad Ilays, the esteemed Figure 1. Chemical structure of Compounds 1 and 2, isolated from
Director of the Botany Department at the University Pistacia chinensis
of Swabi, kindly verified its botanical identity. His
expertise provided a crucial validation step, ensuring the pure crystals of the compounds, ensuring their suitability
20
credibility of the sample used in our study. The specimen, for subsequent chemical and biological analyses. This
meticulously labeled as UOS/Bot-55, has been preserved careful and methodical approach highlights the precision
in the department’s herbarium, serving as a reference for and thoroughness of our efforts to extract and purify the
future research and verification. bioactive components of P. chinensis, paving the way for
their potential application in therapeutic research.
2.2. Extraction and isolation
2.3. Anti-cancer activity
The bark of P. chinensis underwent a meticulous preparation
process to ensure the integrity of the plant material for To evaluate the inhibitory effects of the compounds on cell
further study. Initially, the bark was thoroughly washed growth, the U87 cell line was initially chosen as a model,
with clean water to remove any surface impurities and was providing a reliable framework for assessing cytotoxicity.
then air-dried in a shaded area to prevent any degradation This step allowed us to systematically investigate the
from sunlight. A total of 7.34 kg of shade-dried bark were impact of specific bioactive compounds, identified as
finely ground into a powder using a grinder, yielding Compound 1 and Compound 2, on the suppression of
7.20 kg of powdered material. This finely processed bark cell proliferation. To quantify their cytotoxic potential, we
was then immersed in methanol for 16 days to facilitate an employed the MTT assay, a well-regarded and extensively
exhaustive extraction of its bioactive compounds. validated method for determining cell viability and
measuring cytotoxic effects. This assay enabled a precise
After the 16-day soaking period, the methanolic extract evaluation of the degree of growth inhibition imposed by
was carefully filtered to remove any solid residues and then these compounds on U87 cells, facilitating a comprehensive
concentrated using a rotary evaporator under controlled comparison of their relative efficacy in inhibiting tumor
low temperatures and reduced pressure. This technique cell proliferation. 21
was employed to preserve the chemical integrity of the
extract, resulting in a crude extract weighing 81.29 g. 2.4. In silico analysis
The crude extract was subjected to a fractionation 2.4.1. Retrieval of phytochemicals
process using a separating funnel for further purification. The anti-cancer phytochemicals, identified as Compounds
This approach effectively separated the extract into distinct 1 and 2, underwent an extensive in silico analysis to explore
fractions: hexane (5.01 g), chloroform (17.12 g), ethyl their potential biological activities. Initially, their 2D and
acetate (6.43 g), and methanolic (32.98 g) fractions. The 3D structures were constructed using advanced molecular
methanolic fraction, known for its enriched content of modeling tools. Specifically, ChemDraw Professional 16.0
bioactive compounds, was then analyzed using thin-layer software (version 16.0.1.4.77) was employed to create the
chromatography (TLC) to assess its chemical profile. 2D and 3D conformations using ChemDraw and Chem3D
The methanolic fraction was processed through silica gel applications. These conformations were subsequently
column chromatography to isolate the specific compounds. stored in both structure data file (SDF) and molecular
The column was eluted with a mixture of methanol and (MOL) formats to facilitate further analyses, including
chloroform in a 4:96 ratio, a strategic choice facilitating docking simulations and density functional theory (DFT)
the separation of the targeted compounds. This method evaluations.
led to the successful isolation of two distinct compounds,
Compound 1 and Compound 2. The final purification of 2.4.2. Retrieval of target proteins
Compound 1 and Compound 2 having various functional To identify the protein targets implicated in the anti-cancer
groups (Figure 1), including hydroxyl, ketone, and ether, activity of these compounds, we accessed the Protein
was achieved through recrystallization using a methanol Data Bank (PDB) (RCSB PDB: Homepage) database. Ten
and chloroform solution in a 1:1 ratio. This process yielded crucial protein targets known to play significant roles
Volume 9 Issue 1 (2025) 146 doi: 10.36922/ejmo.5768