Page 157 - EJMO-9-1
P. 157
Eurasian Journal of Medicine and
Oncology
Potential of flavonoids against glioblastoma
effect significantly escalated after 48 h, reaching 60.34% achieving 61.54% inhibition after 48 h (P < 0.01). At the
(P < 0.05). Increasing the concentration to 50 µg/mL led to highest concentration (75 µg/mL), Compound 2 exhibited
an improvement in the 24-h inhibitory activity, recorded the strongest inhibitory effect, with a marked reduction in
at 14.35%, which further amplified to 65.23% at the 48-h cell viability to 30.06% at 24 h, further reaching 68.98%
mark (P < 0.01). At the highest concentration tested after 48 h (P < 0.001).
(75 µg/mL), Compound 1 demonstrated enhanced anti- In general, both compounds demonstrated potent
cancer efficacy, resulting in a 14.98% reduction in viability
after 24 h and increasing to 70.11% after 48 h (P < 0.001). anti-cancer activity against the U87 glioblastoma cell line,
demonstrating increased efficacy with higher concentrations
Compound 2 exhibited a similar pattern of dose- and prolonged exposure times. These findings highlight
dependent inhibition. At 25 µg/mL, the compound the promising potential of these isolated compounds as
displayed an inhibition of 9.54% at 24 h, which significantly therapeutic agents in the treatment of glioblastoma.
increased to 55.09% after 48 h (P < 0.05). The intermediate
concentration of 50 µg/mL induced a more pronounced 3.2. In silico results
effect, reducing cell viability by 16.09% after 24 h and The 2D and 3D structures, as well as the SMILES notations
of the phytochemicals extracted from P. chinensis, are
Table 1. Anti‑cancer activity of compounds isolated from summarized in Table 2. These two bioactive compounds,
Pistacia chinensis in the glioblastoma (U87) cell line designated as Compound 1 and Compound 2, underwent
Sample Dose (µg/mL) Time (h) comprehensive in silico studies to elucidate their molecular
24 48 characteristics and potential bioactivity.
Compound 1 25 10.11 60.34 These conformations and their respective SMILES
50 14.35 65.23 representations provide valuable insights into the molecular
75 14.98 70.11 properties and facilitate further computational analyses,
Compound 2 25 9.54 55.09 including docking studies and ADME evaluations. The
detailed structural information plays a critical role in
50 16.09 61.54 understanding the pharmacological potential of these
75 30.06 68.98 phytochemicals.
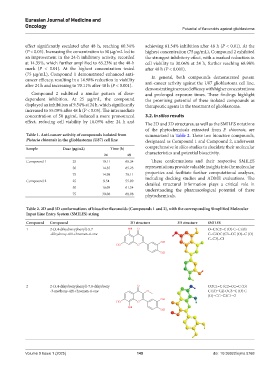
Table 2. 2D and 3D conformations of bioactive flavonoids (Compounds 1 and 2), with the corresponding Simplified Molecular
Input Line Entry System (SMILES) string
Compound Compound 2D structure 3D structure SMILES
1 2-(3,4-dihydroxyphenyl)-5,7 O=C1C2=C (O) C=C (O)
-dihydroxy-4H-chromen-4-one C=C2OC (C3=CC (O)=C (O)
C=C3)=C1
2 2-(3,4-dihydroxyphenyl)-7,8-dihydroxy COC1=C (C2=CC=C (O)
-3-methoxy-4H-chromen-4-one C (O)=C2) OC3=C (O) C
(O)=CC=C3C1=O
Volume 9 Issue 1 (2025) 149 doi: 10.36922/ejmo.5768