Page 212 - EJMO-9-3
P. 212
Eurasian Journal of
Medicine and Oncology FN3K–Nrf2 axis inhibition in breast cancer
accommodates unequal variances and is appropriate homologous templates, highlighting the conserved regions
for datasets with small sample sizes. To mitigate the and secondary structural elements.
51
risk of false positives due to multiple comparisons, the Quality assessment of these 50 templates was performed
Benjamini-Hochberg procedure was applied to control the using QMEAN and GMQE scores. QMEAN evaluates the
False Discovery Rate (FDR). 52 overall geometric and energetic quality of the model, while
2.3.3. Significance thresholds used GMQE estimates the expected accuracy of the homology
model. Templates were ranked based on these criteria,
The following significance thresholds were applied in with the top three templates demonstrating the highest
the analysis: p<0.05 (*; marginally significant), p<0.01 scores and best alignment with the target sequence. In
(**; significant), p<0.001 (***; highly significant), p≥0.05 → the subsequent protein modeling, three homology models
(not significant). All statistical analyses were conducted in (Figure 2) were developed, and the first model, based on
Python utilizing the SciPy and statsmodels libraries, with the template from Arabidopsis thaliana FN3K, exhibited
statsmodels. stats. multitest applied for FDR correction. a sequence similarity of 90.61% with human FN3K and
3. Results and discussion was selected for the study. This model was evaluated using
GMQE and QMEAN scoring functions, where values
3.1. SBVS approaching 1.0 on a scale from 0 to 1 indicate high model
quality. The modeled protein structure (Model-1) achieved
3.1.1. Template selection and model validation satisfactory scores, meeting the quality attributes necessary
Template selection for homology modeling of the human for a reliable model.
FN3K enzyme was carried out by aligning the target protein Further validation was carried out using the
sequence with evolutionarily related structures available PROCHECK tool to assess the stereochemical quality
in the SWISS-MODEL template library. To identify of the model. PROCHECK provided a Ramachandran
suitable homologous templates, the FN3K amino acid plot (Figure 3), which is instrumental in evaluating the
sequence was queried against the PDB-BLAST database. overall geometry of the protein model based on residue-
This search initially yielded 5,643 potential templates that by-residue analysis of backbone dihedral angles (ϕ and ψ).
matched the target sequence. To refine this extensive list, The Ramachandran plot analysis revealed that most of the
a heuristic approach was employed, narrowing it down residues were located in the favored regions, indicating
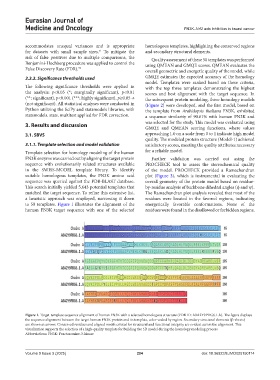
to 50 templates. Figure 1 illustrates the alignment of the energetically favorable conformations. None of the
human FN3K target sequence with one of the selected residues were found in the disallowed or forbidden regions,
Figure 1. Target-template sequence alignment of human FN3K with a selected homologous structure (PDB ID: A0A2Y9PDG6.1.A). The figure displays
the sequence alignment between the target human FN3K protein and its template, color-coded by region. Secondary structural elements (β-sheets)
are shown as arrows. Conserved residues and aligned motifs critical for structural and functional integrity are evident across the alignment. This
visualization supports the selection of a high-quality template for building the 3D model during the homology modeling process
Abbreviations: FN3K: Fructosamine-3-kinase
Volume 9 Issue 3 (2025) 204 doi: 10.36922/EJMO025150114