Page 271 - EJMO-9-3
P. 271
Eurasian Journal of
Medicine and Oncology WGCNA and LASSO for osteoporosis biomarkers
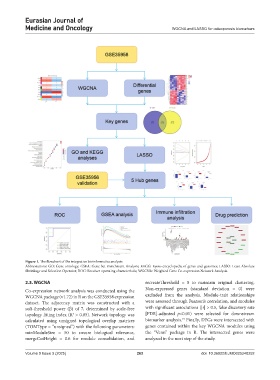
Figure 1. The flowchart of the integrative bioinformatics analysis
Abbreviations: GO: Gene ontology; GSEA: Gene Set Enrichment Analysis; KEGG: Kyoto encyclopedia of genes and genomes; LASSO: Least Absolute
Shrinkage and Selection Operator; ROC: Receiver operating characteristic; WGCNA: Weighted Gene Co-expression Network Analysis.
2.3. WGCNA recreateThreshold = 0 to maintain original clustering.
Co-expression network analysis was conducted using the Non-expressed genes (standard deviation = 0) were
WGCNA package (v1.72) in R on the GSE35958 expression excluded from the analysis. Module-trait relationships
dataset. The adjacency matrix was constructed with a were assessed through Pearson’s correlation, and modules
soft-threshold power (β) of 7, determined by scale-free with significant associations (|r| > 0.5, false discovery rate
topology fitting index (R > 0.85). Network topology was [FDR]-adjusted p<0.01) were selected for downstream
2
15
calculated using unsigned topological overlap matrices biomarker analysis. Finally, DEGs were intersected with
(TOMType = “unsigned”) with the following parameters: genes contained within the key WGCNA modules using
minModuleSize = 50 to ensure biological relevance, the “Venn” package in R. The intersected genes were
mergeCutHeight = 0.6 for module consolidation, and analyzed in the next step of the study.
Volume 9 Issue 3 (2025) 263 doi: 10.36922/EJMO025240252