Page 65 - GPD-3-1
P. 65
Gene & Protein in Disease In silico application of the CoM method
Figure 1. Visualization of the proposed method.
water model, having placed them and centered into a cubic
box at 1 nm minimum distance from the edge of the box.
16
Brooks et al. showed that Charmm27 all-atom force field
was used for the purpose of simulation. Totally, 25 water
+
molecules were substituted with 25 Na ions to bring up
the systems to the neutral net charge. The systems were
relaxed and optimized within-self applying the steepest
descent energy minimization algorithm, until potential
17
energy E < −10 kJmol .
5
−1
pot
The purpose of 100-ps NVT equilibrium phase,
controlled by V-rescale thermostat, was to bring the systems
under the desired temperature of 310 K. V-rescale belongs
to a sophisticated group of algorithms named thermostats
and its role is to maintain a constant temperature level
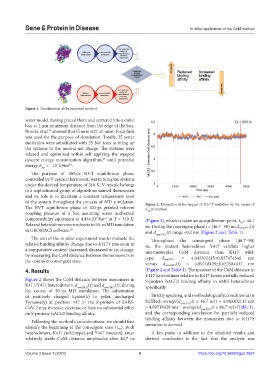
in the system throughout the process of MD simulation. Figure 2. Evaluation of the impact of K417Y mutation by the means of
The NVT equilibrium phase of 100-ps granted referent d (t) method.
coupling pressure of 1 bar, assuming water isothermal com
−1
−5
compressibility equivalent to 4.45×10 bar at T = 310 K. (Figure 2), which is taken an as equilibrium point, t eq.= 46.7
Relaxed heterodimers were subjects to 50-ns MD simulation ns. During the convergent phase t = [46.7−50] ns, d com,K417(t)
in GROMACS software. 18 and d (t) range <0.2 nm (Figure 2 and Table 1).
com,Y417
The aim of the in silico experiment was to evaluate the Throughout the convergent phase [46.7−50]
relative binding affinity change due to K417Y mutation in ns, the mutant heterodimer Y417 exhibits higher
a comparative context: increased, decreased or no change, intermolecular CoM distance than K417 wild-
by measuring the CoM distance between the monomers in type: = 4.943302115±0.037474346 nm
the common convergent state. d com,K417
versus d com,K417(t) = 4.89718429±0.033584437 nm
4. Results (Figure 2 and Table 1). The increase of the CoM distance in
Y417 heterodimer relative to K417 favors partially reduced
Figure 2 shows the CoM distance between monomers in S-protein–hACE2 binding affinity in 6M0J heterodimer
K417/Y417 heterodimers: d com,K417 (t) and d com,Y417 (t), during specifically.
the course of 50-ns MD simulation. The substitution
of positively charged Lysine(k) to polar, uncharged Strictly speaking, and methodologically, condition (a) is
Tyrosine(y) at position 417 in the S-protein of SARS- fulfilled: average(d com,Y417 (t ≥ 46.7 ns)) = 4.943302115 nm
CoV-2 may increase, decrease or have no substantial effect > 4.89718429 nm = average(d com,K417 (t ≥ 46.7 ns)) (Table 1),
on S-protein–hACE2 binding affinity. and the corresponding conclusion for partially reduced
binding affinity between the monomers due to K417Y
Following the method’s considerations, we should first
identify the beginning of the convergent state (t eq.). Both mutation is derived.
heterodimers, K417 (wild-type) and Y417 (mutant), enter A key point in addition to the obtained results and
relatively stable CoM distance amplitudes after 46.7 ns derived conclusion is the fact that the analysis was
Volume 3 Issue 1 (2024) 4 https://doi.org/10.36922/gpd.2657