Page 66 - GPD-3-1
P. 66
Gene & Protein in Disease In silico application of the CoM method
performed during the common convergent state in both major point of interest, which has been analyzed in terms
heterodimers and not throughout the course of the entire of 50-ns MD simulation (Figure 4 and Table 2).
simulation [0 – 50] ns, resembling the protocols in previous In K417 heterodimer, the salt bridge was formed
studies, potentially resulting in misleading conclusion between deprotonated carboxylic acid COO in D30
2,3
−
related to the binding affinity change.
(aspartic acid, hACE2) and the positively charged ε-amino
+
5. Discussion group NH in K417 (Lysine, S-protein) (Figure 4). The
3
salt bridge was changed to hydrogen bond in Y417 mutant,
Non-covalent interactions, specifically involving K417/ formed between D30 carboxylate ion and K417 phenolic
Y417 residues, were analyzed, based on the relaxed, hydroxyl group (-OH) (Figure 4).
crystal pose K417/Y417 PDB structures, using the Ring
3.0 server (https://ring.biocomputingup.it/), for the Although the salt bridge is a k-fold stronger interaction
19
following cutoff values: maximum ionic bond distance than the hydrogen bond, it has been inspected for the
4 Å, maximum hydrogen bond donor-acceptor distance occupancy of these interactions, as the overall impact of a
3.5 Å, maximum π−π stacking distance 4 Å, and Van der strong but temporary interaction may be outcompeted by
Walls radius intersection fraction of <0.01 Å. a weaker but permanent interaction(s).
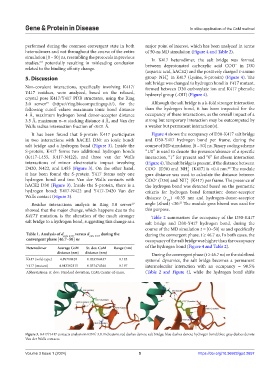
It has been found that S-protein K417 participates Figure 4 shows the occupancy of D30-K417 salt bridge
in two interactions with hACE2 D30: an ionic bond/ and D30-Y417 hydrogen bond per frame, during the
salt bridge and a hydrogen bond (Figure 3). Inside the course of MD simulation [0 – 50] ns. Binary coding scheme
S-protein, K417 forms two additional hydrogen bonds “1/0” is used to denote the presence/absence of a specific
(K417-L455, K417-N422), and three van der Walls interaction, “1” for present and “0” for absent interaction
interactions of minor electrostatic impact involving (Figure 4). The salt bridge is present, if the distance between
D420, N422, and L455 (Figure 3). On the other hand, COO (D30) and NH (K417) is <0.4 nm. The module
+
−
20
3
it has been found the S-protein Y417 forms only one gmx distance was used to calculate the distance between
hydrogen bond and two Van der Walls contacts with COO (D30) and NH (K417) per frame. The presence of
+
−
3
hACE2 D30 (Figure 3). Inside the S-protein, there is a the hydrogen bond was detected based on the geometric
hydrogen bond: Y417-N422 and Y417-D420 Van der criteria for hydrogen bond formation: donor-acceptor
Walls contact (Figure 3). distance (r ) <0.35 nm and hydrogen-donor-acceptor
DA
21
Residue interactions analysis in Ring 3.0 server angle (∡had) <30. The module gmx hbond was used for
19
showed that the major change, which happens due to the this purpose.
K417Y mutation, is the alteration of the much stronger Table 2 summarizes the occupancy of the D30-K417
salt bridge to a hydrogen bond, suggesting this change as a salt bridge and D30-Y417 hydrogen bond, during the
course of the MD simulation t = [0−50] ns and specifically
Table 1. Analysis of d com, K417 versus d com, Y417 during the during the convergent phase, t ≥ 46.7 ns. In both cases, the
convergent phase [46.7−50] ns occupancy of the salt bridge was higher than the occupancy
Heterodimer Average CoM St. dev. CoM Range (nm) of the hydrogen bond (Figure 4 and Table 2).
distance (nm) distance (nm) During the convergent phase (t ≥ 46.7 ns) or the stabilized
K417 (wild-type) 4.89718429 0.033584437 0.185 systems’ dynamics, the salt bridge becomes a permanent
Y417 (mutant) 4.943302115 0.037474346 0.197 intermolecular interaction with an occupancy = 98.5%
Abbreviations: st. dev.: Standard deviation; CoM: Center-of-mass. (Table 2 and Figure 4), while the hydrogen bond shifts
Figure 3. K417/Y417 contacts analysis in RING 3.0. Indicators: red dashes denote salt bridge; blue dashes denote hydrogen bond; blue-gray dashes denote
Van der Walls contacts.
Volume 3 Issue 1 (2024) 5 https://doi.org/10.36922/gpd.2657