Page 29 - GTM-3-3
P. 29
Global Translational Medicine Computational advances in cancer liquid biopsy
Nevertheless, collecting CTCs from a patient’s blood
is not a straightforward task and is strongly influenced by
the cancer type, sampling site, timing, and many clinical
factors such as tumor grade, invasiveness, and size.
Moreover, technical issues that can affect the outcome of
the analysis include marker-dependent or independent
selection and RNA quantity and quality.
Many single-cell RNA-seq analyses of CTCs utilize
antibody staining of epithelial markers for CTC detection,
resulting in a partial/biased sampling of the circulating
cells and upregulated epithelial gene expression in CTCs
compared to controls. Conversely, many CTCs present a
hybrid epithelial/mesenchymal phenotype or are in EMT,
81
as demonstrated by a recent paper on metastatic gastric
cancer, where the authors combined size-dependent
82
recovery of CTCs with CD45-negative selection, allowing
the inclusion of EMT-induced CTCs. On the other hand,
epithelial non-tumor cells are shed in blood during
inflammation, infections, and invasive procedures such
as surgery and biopsies, which may be confounded for
CTCs in cancer patients, patients with benign tumors, and
healthy individuals. These issues must be considered as the
detection of cancer in asymptomatic patients, which is the
primary goal of detection tests, requires high specificity.
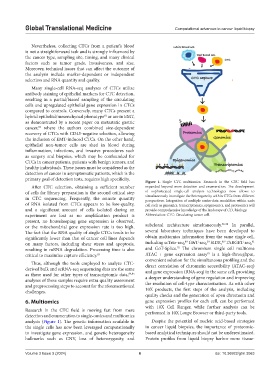
Figure 1. Single CTC multiomics. Research in the CTC field has
After CTC selection, obtaining a sufficient number expanded beyond mere detection and enumeration. The development
of cells for library preparation is the second critical step of sophisticated single-cell analysis technologies now allows to
in CTC sequencing. Frequently, the minute quantity simultaneously investigate the heterogeneity within CTCs from different
perspectives. Integration of multiple omics-data modalities within each
of RNA isolated from CTCs appears to be low-quality, cell such as genomics, transcriptomics, epigenomics, and proteomics will
and a significant amount of cells isolated during an provide comprehensive knowledge of the landscape of CTC biology.
experiment are lost as no amplification product is Abbreviation: CTC: Circulating tumor cell.
present, no housekeeping gene expression is observed,
or the mitochondrial gene expression rate is too high. subclonal architecture simultaneously. 86-88 In parallel,
The fact that the RNA quality of single CTCs tends to be several laboratory techniques have been developed to
significantly lower than that of cancer cell lines depends obtain multiomics information from the same single cell,
90
91
89
59
on many factors, including shear stress and apoptosis, including scTrio-seq, G&T-seq, SIDR, TARGET-seq,
92
resulting in mRNA degradation. Processing time is also and GoT-Splice. The chromium single cell multiome
93
critical to maximize capture efficiency. 83 ATAC + gene expression assay is a high-throughput,
convenient solution for the simultaneous profiling and the
Thus, although the tools employed to analyze CTC- direct correlation of chromatin accessibility (ATAC-seq)
derived bulk and scRNA-seq sequencing data are the same and gene expression (RNA-seq) in the same cell, providing
as those used for other types of transcriptomic data, 84,85 a deeper understanding of gene regulation and improving
analyses of these samples require extra quality assessment
and preprocessing steps to account for the aforementioned the resolution of cell-type characterization. As with other
challenges. 10X products, the first steps of the analysis, including
quality checks and the generation of open chromatin and
6. Multiomics gene expression profiles for each cell, can be performed
with 10X Cell Ranger, while further analysis can be
Research in the CTC field is moving fast from mere performed in 10X Loupe Browser or third-party tools.
detection and enumeration to single-omics and multiomics
analysis (Figure 1). The genetic information available in Despite the potential of nucleic acid-based strategies
the single cells has now been leveraged computationally in cancer liquid biopsies, the importance of proteomic-
to investigate gene expression, and genetic heterogeneity based analytical techniques should not be underestimated.
hallmarks such as CNV, loss of heterozygosity, and Protein profiles from liquid biopsy harbor more tissue-
Volume 3 Issue 3 (2024) 6 doi: 10.36922/gtm.3063