Page 15 - GTM-4-2
P. 15
Global Translational Medicine Game-changing drug response prediction
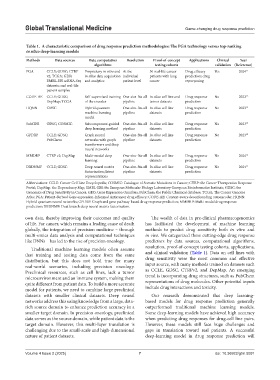
Table 1. A characteristic comparison of drug response prediction methodologies: The PGA technology versus top-ranking
in silico deep-learning models
Methods Data sources Data computation Resolution Proof-of-concept Applications Clinical Year
algorithms testing cohorts validation (Reference)
PGA CCLE; GDSC; CTRP Proprietary in vitro and At the 30 real-life cancer Drug efficacy Yes 2024 11
v2; TCGA; GEO; in silico data acquisition individual patients with lung prediction; drug
EMBL-EBI scRNA-Seq and analytics patient level cancer repurposing
datasets; and real-life
patient samples
CODE-AE CCLE; GDSC; Self-supervised training One-size-fits-all In silico cell line and Drug response No 2022 12
DepMap; TCGA of the encoder pipeline tumor datasets prediction
HQNN GDSC Hybrid quantum One-size-fits-all In silico cell line Drug response No 2023 13
machine learning pipeline datasets prediction
model
SubCDR GDSC; COSMIC Subcomponent-guided One-size-fits-all In silico cell line Drug response No 2023 14
deep learning method pipeline datasets prediction
GPDRP CCLE; GDSC; Graph neural One-size-fits-all In silico cell line Drug response No 2023 15
PubChem networks with graph pipeline datasets prediction
transformers and deep
neural networks
MMDRP CTRP v2; DepMap Multi-modal deep One-size-fits-all In silico cell line Drug response No 2024 16
learning pipeline datasets prediction
DBDNMF CCLE; GDSC Deep neural matrix One-size-fits-all In silico cell line Drug response No 2024 17
factorization; latent pipeline datasets prediction
representations
Abbreviations: CCLE: Cancer Cell Line Encyclopedia; COSMIC: Catalogue of Somatic Mutations in Cancer; CTRP: the Cancer Therapeutics Response
Portal; DepMap: the Dependency Map; EMBL-EBI: the European Molecular Biology Laboratory-European Bioinformatics Institute; GDSC: the
Genomics of Drug Sensitivity in Cancer; GEO: Gene Expression Omnibus; PubChem: the Public Chemical database; TCGA: The Cancer Genome
Atlas; PGA: Patient-derived Gene expression-informed Anticancer drug efficacy; CODE-AE: Context-aware deconfounding autoencoder, HQNN:
Hybrid quantum neural networks; GPDRP: Graph and gene pathway-based drug response prediction; MMDRP: Multi-modal drug response
prediction; DBDNMF: Dual branch deep neural matrix factorization.
own data, thereby improving their outcomes and quality The wealth of data in pre-clinical pharmacogenomics
of life. For cancer, which remains a leading cause of death has facilitated the development of machine learning
globally, the integration of precision medicine – through methods to predict drug sensitivity both in vitro and
multi-omics data analysis and computational techniques in vivo. We categorized these cutting-edge drug response
like DNNs – has led to the rise of precision oncology. predictors by data sources, computational algorithms,
Traditional machine learning models often assume resolution, proof-of-concept testing cohorts, applications,
that training and testing data come from the same and clinical validation (Table 1). Data on cell lines with
distribution, but this does not hold true for many drug sensitivity were the most common and effective
real-world scenarios, including precision oncology. input source, with many methods trained on datasets such
Preclinical resources, such as cell lines, lack a tumor as CCLE, GDSC, CTRPv2, and DepMap. An emerging
microenvironment and an immune system, making them trend is incorporating drug structures, such as PubChem
quite different from patient data. To build a more accurate representations of drug molecules. Other potential inputs
model for patients, we need to combine large preclinical include drug interactions and toxicity.
datasets with smaller clinical datasets. Deep neural Our research demonstrated that deep learning-
networks address this using knowledge from a large, data- based models for drug response prediction generally
rich source domain to enhance prediction accuracy in a outperformed traditional machine learning models.
smaller target domain. In precision oncology, preclinical Some deep-learning models have achieved high accuracy
data serves as the source domain, while patient data is the when predicting drug responses for drug-cell line pairs.
target domain. However, this multi-layer translation is However, these models still face huge challenges and
challenging due to the small-scale and high-dimensional gaps in translation toward real patients. A successful
nature of patient datasets. deep-learning model in drug response prediction will
Volume 4 Issue 2 (2025) 7 doi: 10.36922/gtm.5091