Page 16 - GTM-4-2
P. 16
Global Translational Medicine Game-changing drug response prediction
likely depend on the effective combination of real-world individually detect and identify actionable biomarkers to
real-time data with learned representations and carefully benefit only the responders. The urgent need now is for a
selected manual features. It is, therefore, crucial to harness trainable platform that can quickly and accurately predict
the potential of deep learning-based models before drug efficacy for those patient non-responders who have
conducting clinical studies. been excluded from precision medicine.
In summary, deep-learning models often function PGA technology is the world-first gene-to-drug
as black-box models, needing more human interfaces, platform, offering high-value-added diagnostic and
which make decision-making challenging. It is hard to fix therapeutic solutions. For drug response prediction, PGA
11
variations in biology that show high-degree of heterogeneity combines the high-resolution cell-free messenger RNA
between patients, tumors, and their microenvironment. (mRNA) profiling with gene expression mapping dedicated
Future researches should incorporate strategies for to identify a patient-unique signature. Based on this in vitro
enhancing their generalizability and interpretability. configuration, combining in silico digital curation, data
fusion, and computation ensures accurate identification
7. A gene-to-drug renaissance of potential hits (after screening of more than 700 Food
Precision oncology is the use of omics data to tailor and Drug Administration-approved, clinical trial, and
therapy for an individual cancer patient; however, only investigational drugs). This unprecedented combination of
10% of patients actually benefit from precision therapy in vitro and in silico approaches allows PGA to effectively
today. 18,19 Improving drug response prediction in the rest map and identify effective drugs at the individual patient
90% non-responders will significantly benefit many more level, providing a significant boost to improve treatment
cancer patients. It is no longer viable for companies to outcome in precision oncology, especially for those patients
load up their instruments with bespoke capabilities that with limited treatment options (Figure 3).
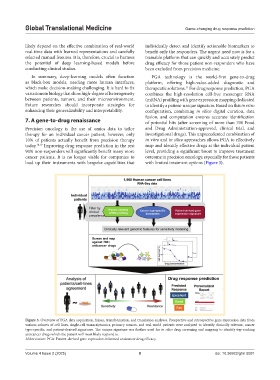
Figure 3. Overview of PGA data acquisition, fusion, transformation, and translation analyses. Prospective and retrospective gene expression data from
various cohorts of cell lines, single-cell transcriptomics, primary tumors, and real-world patients were analyzed to identify clinically relevant, cancer
type-specific, and patient-derived signatures. The unique signature was further used for in silico drug screening and mapping to identify top-ranking
anticancer drugs which the patient will most likely respond to.
Abbreviation: PGA: Patient-derived gene expression-informed anticancer drug efficacy.
Volume 4 Issue 2 (2025) 8 doi: 10.36922/gtm.5091