Page 63 - IJB-9-4
P. 63
International Journal of Bioprinting Machine learning and 3D bioprinting
after deposition. ML is expected to optimize process-
related parameters, such as the type of crosslinking and the
density of the formed crosslinks, and to explore the trade-
off between the stiffness of the deposited structures and
the printability of hydrogel inks. However, to date, no such
studies have been reported.
It is often assumed that ML methods can accelerate
biomaterial/bioink studies by optimizing formulations,
viscosity, and the consequent rheological properties to
control the biological and mechanical performance. The
integration of multiple properties can also be considered
as the evaluation metrics for the optimization tasks. To
develop a stable Col-I bioink which is a mixture of 15
compositions , the bioink concentration should be
[43]
optimized in terms of both mechanical properties and
transparency. Such research would significantly advance
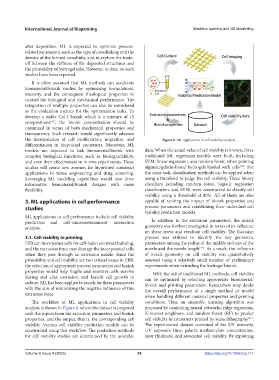
the manipulation of cell proliferation, migration, and Figure 6. ML application in cell viability analysis.
differentiation in bioprinted constructs. Moreover, ML
models are expected to link biomaterial/bioink with data. When the actual value of cell viability is known, three
complex biological functions, such as biodegradability, traditional ML regression models were built, including
and even their effectiveness in in vivo experiments. These SVM, linear regression, and random forest, when printing
studies will create new avenues for bioprinted construct alginate/gelatin-based hydrogels loaded with cells . For
[34]
applications in tissue engineering and drug screening. the same task, classification methods can be applied when
Leveraging ML modeling capabilities would also drive using a threshold to judge the cell viability. Three binary
informative biomaterial/bioink designs with more classifiers including random forest, logistic regression
flexibility. classification, and SVM, were constructed to classify cell
viability using a threshold of 80%. All of them are also
3. ML applications in cell performance capable of ranking the impact of bioink properties and
studies process parameters and establishing their individual cell
viability prediction models.
ML applications in cell performance include cell viability
prediction and cell–microenvironment interaction In addition to the extrusion parameters, the nozzle
analysis. geometry was further investigated in terms of its influence
on shear stress and resultant cell viability. The Gaussian
3.1. Cell viability in printing process was utilized to identify the key geometric
EBB can incorporate cells for cell-laden construct building, parameters among the radius of the middle and exit of the
and the extrusion force may damage the incorporated cells nozzle and the nozzle length . As a result, the influence
[44]
when they pass through an extrusion nozzle. Since the of nozzle geometry on cell viability was quantitatively
printability and cell viability are two critical issues in EBB, assessed using a relatively small number of preliminary
the selection of appropriate process parameters and bioink experiments when extruding the hydrogel bioink.
properties would help fragile and sensitive cells survive With the aid of traditional ML methods, cell viability
during and after extrusion, and benefit cell growth in can be optimized by selecting appropriate biomaterial/
culture. ML has been applied to search for these parameters bioink and printing parameters. Researchers may doubt
with the aim of minimizing the negative influence of this the overall performance of a single method or model
extrusion force. when handling different material properties and printing
The workflow of ML applications in cell viability conditions. Thus, an ensemble learning algorithm was
analysis is shown in Figure 6, where the dataset is prepared proposed by combining neural networks, ridge regression,
with the inputs from the extrusion parameters and bioink K-nearest neighbors, and random forest (RF) to predict
properties, and the output, that is, the corresponding cell cell viability in constructs printed by stereolithography .
[45]
viability. Various cell viability prediction models can be The experimental dataset consisted of the UV intensity,
constructed using this workflow. The prediction methods UV exposure time, gelatin methacrylate concentration,
for cell viability studies are determined by the available layer thickness, and associated cell viability. By exploiting
Volume 9 Issue 4 (2023) 55 https://doi.org/10.18063/ijb.717