Page 65 - IJB-9-4
P. 65
International Journal of Bioprinting Machine learning and 3D bioprinting
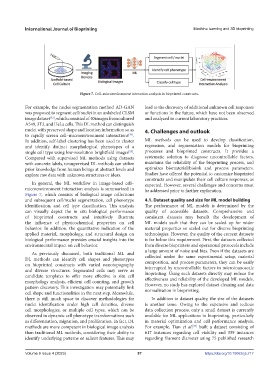
Figure 7. Cell–microenvironment interaction analysis in bioprinted constructs.
For example, the nuclei segmentation method AD-GAN lead to the discovery of additional unknown cell responses
was proposed to segment cell/nuclei in an unlabeled CLSM or functions in the future, which have not been observed
image dataset , which consisted of 40 images from cultured and analyzed in current laboratory practices.
[52]
A549, 3T3, and HeLa cells. This DL method can distinguish
nuclei with preserved shape and location information so as 4. Challenges and outlook
to rapidly screen cell–microenvironment interactions .
[36]
In addition, self-label clustering has been used to cluster ML methods can be used to develop classification,
and identify distinct morphological phenotypes of a regression, and segmentation models for bioprinting
single cell type using low-resolution brightfield images . processes and bioprinted constructs. It provides a
[53]
Compared with supervised ML methods using datasets systematic solution to diagnose uncontrollable factors,
with concrete labels, unsupervised DL methods can utilize maintains the reliability of the bioprinting process, and
prior knowledge from human beings at abstract levels and optimizes biomaterial/bioink and process parameters.
explore raw data with unknown structures or ideas. Studies have offered the potential to customize bioprinted
constructs and manipulate their cell culture responses, as
In general, the ML workflow in image-based cell– expected. However, several challenges and concerns must
microenvironment interaction analysis is summarized in be addressed prior to further exploration.
Figure 7, which consists of biological image collections
and subsequent cell/nuclei segmentation, cell phenotype 4.1. Dataset quality and size for ML model building
identification, and cell type classification. This analysis The performance of ML models is determined by the
can visually depict the in situ biological performance quality of accessible datasets. Comprehensive and
of bioprinted constructs and intuitively illustrate consistent datasets may benefit the development of
the influence of physicochemical properties on cell ML models such that they can be scaled up for wider
behavior. In addition, the quantitative indication of the material properties or scaled out for diverse bioprinting
applied material, morphology, and structural design on technologies. However, the quality of the current datasets
biological performance provides crucial insights into the is far below this requirement. First, the datasets collected
environmental impact on cell behavior. from diverse bioprinters and operational protocols include
a large amount of noise and bias. Even if the datasets are
As previously discussed, both traditional ML and
DL methods can identify cell shapes and phenotypes collected under the same experimental setup, material
composition, and process parameters, they can be easily
on bioprinted constructs with varied nanotopography interrupted by uncontrollable factors in micro/nanoscale
and diverse structures. Segmented cells may serve as bioprinting. Using such datasets directly may reduce the
candidate templates to offer more effective in situ cell effectiveness and reliability of the developed ML models.
morphology analysis, efficient cell counting, and growth However, no study has explored dataset cleaning and data
pattern discovery. This investigation may potentially link normalization in bioprinting.
cell shape and functionalities in the next step. Meanwhile,
there is still much space to discover methodologies for In addition to dataset quality, the size of the datasets
nuclei identification under high cell densities, diverse is another issue. Owing to the expensive and tedious
cell morphologies, or multiple cell types, which can be data collection process, only a small dataset is currently
observed in dynamic cell phenotype transformations such available for ML applications in bioprinting, particularly
as differentiation, migration, and proliferation. In fact, DL in material optimization and cell performance analysis.
methods are more competent in biological image analysis For example, Tian et al. built a dataset consisting of
[34]
than traditional ML methods, considering their ability to 617 instances regarding cell viability and 339 instances
identify underlying patterns or salient features. This may regarding filament diameter using 75 published research
Volume 9 Issue 4 (2023) 57 https://doi.org/10.18063/ijb.717