Page 24 - ITPS-8-1
P. 24
INNOSC Theranostics and
Pharmacological Sciences Transcriptome-based RNA sequencing
References 17 18 19 20 (Cont'd...)
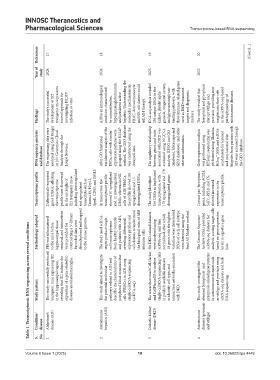
Year of study 2024 2024 2023 2022
The study’s successful development of IE2 transgenic mice presents a novel approach for investigating HCMV infection in vitro. AIH is an immunological condition characterized by autoantibodies, hypergammaglobulinemia, and persistent active hepatitis. Understanding the molecular mechanisms in PBMC-derived monocyte and NK cell clusters could aid AIH therapy. RNA-seq analysis revealed 356 unique DEGs in the kidney, platele
Findings markers.
RNA sequence analysis and database The sequencing data were analyzed using Cell Ranger or 10× Genomics Cloud Analysis through the Loupe Browser. After GO functional enrichment analysis of the DEGs, cells with similar expression patterns were grouped using the UMAP method. The GEO database was constructed using the obtained data. The regulatory relationship between genes and traits associated with DKD was evaluated using WGCNA
Transcriptome profile Differentially expressed genes (DEGs) affecting the transcriptome profile were discovered in the microglia of IE2 transgenic mice, including downregulated and upregulated transcripts such as Tmem119, P2ry12, ApoE, CD83, and ISG15. In monocytes, the transcriptome profile showed 87 upregulated and 12 downregulated DEGs, whereas in NK cells of AIH PBMCs, the profile showed 101 upregulated and 15 dow
Methodology adopted PBS was administered to the mice before hippocampal tissue removal, and instructions were provided for constructing a 3ʹ-library. Barcoded data were demultiplexed and mapped to the mouse genome. The study used scRNA- seq to produce single- cell transcriptomes from healthy controls and patients with AIH, revealing differential expression profiles and cell clusters to understand the biological roles of AIH
Table 1. Transcriptomic RNA sequencing across several conditions
This study successfully produced transgenic mice expressing IE2 in the hippocampal region, revealing that IE2 induces the expression of a gene related to disease-associated microglia. This study aimed to investigate pathways related to AIH and describe the characteristics of peripheral blood mononuclear cells (PBMCs) in AIH using single-cell RNA sequencing The researchers used CellMarker and CellPhoneDB to analyze single-cell RNA seq
Study pattern the potential molecular (scRNA-seq). with DKD. RNA sequencing.
Condition/ disease Alzheimer’s disease (AD) Autoimmune hepatitis (AIH) Diabetic kidney disease (DKD) Autoimmune diseases (psoriasis and vitiligo)
S. No. 1 2 3 4
Volume 8 Issue 1 (2025) 18 doi: 10.36922/itps.4449