Page 90 - MI-1-2
P. 90
Microbes & Immunity Identifying hydrogenase orthologs in the human proteome
identify regions of similarity between complex I subunits A B
and [NiFe] hydrogenases. 35
2.3. Clustal Ω
Prokaryotic sequences displaying similarities with the
human proteins NDUFS2 and NDUFS7 were then aligned
and analyzed using the Clustal Ω program. 36
2.4. ScanProsite C D
Using selected motifs of interest, an analysis was performed
with ScanProsite to identify predicted sites of post-
translational modifications, including N-glycosylation,
N-myristoylation, and phosphorylation, known to support
pro-oxidative and proinflammatory signaling pathways.
This investigation focused on pinpointing redox-sensitive
sites that could potentially facilitate H₂-driven antioxidant
and anti-inflammatory signaling activity. 37
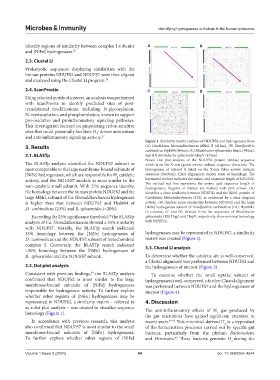
Figure 1. Similarity matrix analyses of NDUFS2 and hydrogenases from
3. Results (A) Candidatus Heimdallarchaeota MbhL B (418aa), (B) Desulfovibrio
carbinolicus Hyd494 (494aa), (C) Rhodobacter sphaeroides HupL (596aa),
3.1. BLASTp and (D) Rhodobacter sphaeroides HupV (475aa)
Notes: Dot plot analysis of the NDUFS2 protein (463aa) sequence
The BLASTp analysis identified the NDUFS2 subunit as which is on the X-axis (green arrows indicate sequence direction). The
most comparable to the large membrane-bound subunits of hydrogenase of interest is listed on the Y-axis (blue arrows indicate
[NiFe] hydrogenases, which are responsible for H catalytic sequence direction). Clear alignments denote areas of homology. The
2
activity, and the NDUFS7 module as most similar to the horizontal red line indicates the amino acid sequence length of NDUFS2.
The vertical red line represents the amino acid sequence length of
non-catalytic small subunit. With 27% sequence identity, hydrogenases. Regions of interest are marked with pink arrows. (A)
the homology between the human protein NDUFS2 and the identifies a close similarity between NDUFS2 and the MbhL protein of
large MbhL subunit of Ca. Heimdallarchaeota hydrogenase Candidatus Heimdallarchaeota (CH), as evidenced by a clear diagonal
is higher than that between NDUFS2 and Hyd494 of pattern. (B) displays some similarities between NDUFS2 and the large
D. carbinolicus (23%) and R. sphaeroides (<20%). [NiFe] hydrogenase subunit of Desulfovibrio carbinolicus (DC; Hyd494).
In contrast, (C and D), derived from the sequences of Rhodobacter
Exceeding the 25% significance threshold, the BLASTp sphaeroides (RS) HupL and HupV, respectively, show minimal homology
38
analysis of Ca. Heimdallarchaeota showed a 35% similarity with NDUFS2.
with NDUFS7. Notably, the BLASTp search indicated
35% homology between the [NiFe] hydrogenases of hydrogenases may be represented in NDUFS7, a similarity
D. carbinolicus and the NDUFS7 subunit of mitochondrial matrix was created (Figure 2).
complex I. Conversely, the BLASTp search indicated 3.3. Clustal Ω analysis
<20% homology between the [NiFe] hydrogenases of
R. sphaeroides and the NDUFS7 subunit. To determine whether the catalytic site is well-conserved,
a Clustal alignment was performed between NDUFS2 and
3.2. Dot plot analysis the hydrogenases of interest (Figure 3).
Consistent with previous findings, the BLASTp analysis To examine whether the small uptake subunit of
29
confirmed that NDUFS2 is most similar to the large hydrogenases is well-conserved, a further Clustal alignment
membrane-bound subunits of [NiFe] hydrogenases was performed between NDUFS7 and the hydrogenases of
responsible for hydrogenase activity. To further explore interest (Figure 4).
whether other regions of [NiFe] hydrogenases may be
represented in NDUFS2, a similarity matrix – referred to 4. Discussion
as a dot plot analysis – was created to visualize sequence The anti-inflammatory effects of H gas produced by
homology (Figure 1). 2
the gut microbiota have gained significant attention in
In accordance with previous research, this analysis recent years. 39-42 This microbial-derived H is a byproduct
2
also confirmed that NDUFS7 is most similar to the small of the fermentation processes carried out by specific gut
membrane-bound subunits of [NiFe] hydrogenases. bacteria, particularly from the phylum Bacteriodetes
To further explore whether other regions of [NiFe] and Firmicutes. These bacteria generate H during the
43
2
Volume 1 Issue 2 (2024) 84 doi: 10.36922/mi.4544