Page 91 - MI-1-2
P. 91
Microbes & Immunity Identifying hydrogenase orthologs in the human proteome
anaerobic metabolism of dietary fibers that are indigestible methanogenic archaea, sulfate-reducing bacteria, and
by the human body. H produced by commensal bacteria acetogenic bacteria, which employ H as an electron donor
2
2
can be utilized by other microbial populations, particularly in their respective metabolic pathways. This symbiotic
8
relationship between H producers and consumers helps
2
A B maintain a balanced intestinal environment. Imbalances,
such as an overproduction of H , can lead to gastrointestinal
2
disturbances like small intestine bacterial overgrowth.
44
In contrast, insufficient H utilization, which is a more
2
common occurrence, may impair microbial metabolic
efficiency, leading to an increase in ROS and subsequent
inflammation.
C D The gut microbiome is well established as a modulator
of systemic inflammation through various mechanisms,
45
including the regulation of immune cell activity,
cytokine production, and the maintenance of intestinal
46
epithelial integrity. A healthy microbiome, enriched in
47
H -utilizing microbes, is critical for preventing leaky gut
2
syndrome, which can lead to systemic inflammation by
allowing bacterial endotoxins to enter the bloodstream.
48
Furthermore, microbial-produced H has been shown to
2
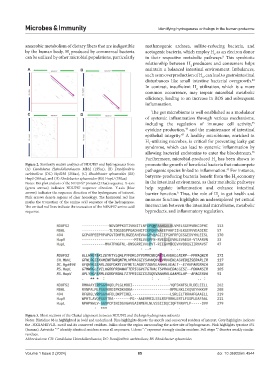
Figure 2. Similarity matrix analyses of NDUFS7 and hydrogenases from promote the growth of beneficial bacteria that outcompete
(A) Candidatus Heimdallarchaeota MbhJ (155aa), (B) Desulfovibrio pathogenic species linked to inflammation. For instance,
43
carbinolicus (DC) Hyd258 (258aa), (C) Rhodobacter sphaeroides (RS) butyrate-producing bacteria benefit from the H economy
HupS (369aa), and (D) Rhodobacter sphaeroides (RS) HupU (330aa) 2
Notes: Dot plot analysis of the NDUFS7 protein (213aa) sequence. X-axis in the intestinal environment, as their metabolic pathways
(green arrows) indicates NDUFS7 sequence direction. Y-axis (blue help regulate inflammation and enhance intestinal
arrows) indicates the sequence direction of the hydrogenase of interest. barrier function. Thus, the role of H in gut health and
8
2
Pink arrows denote regions of clear homology. The horizontal red line immune function highlights an underexplored yet critical
marks the truncation of the amino acid sequence of the hydrogenase.
The vertical red lines indicate the truncation of the NDUFS7 amino acid intersection between the intestinal microbiome, metabolic
sequence. byproducts, and inflammatory regulation.
Figure 3. Short sections of the Clustal alignment between NDUFS2 and the large hydrogenase subunits
Notes: Histidine 88 is highlighted in bold and underlined. Blue highlights denote the motifs and conserved residues of interest. Grey highlights indicate
the -HXXAHXVLR- motif and its conserved residues. Italics show the region surrounding the active site of hydrogenases. Pink highlights tyrosine 151
(human). Asterisks “*” identify identical residues across all sequences. Colons “:” represent strongly similar residues. Full stops “.” denotes weakly similar
residues.
Abbreviations: CH: Candidatus Heimdallarchaeota; DC: Desulfovibrio carbinolicus; RS: Rhodobacter sphaeroides.
Volume 1 Issue 2 (2024) 85 doi: 10.36922/mi.4544