Page 102 - MI-2-4
P. 102
Microbes & Immunity Brachyspira pilosicoli novel outer membrane proteins
A B C D E
G H I J
F
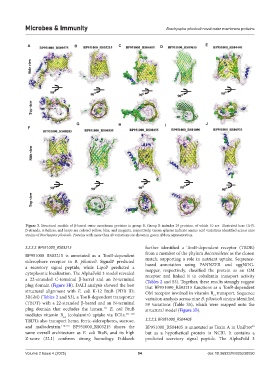
Figure 3. Structural models of β-barrel outer membrane proteins in group B. Group B includes 29 proteins, of which 10 are illustrated here (A-J).
β-strands, α-helices, and loops are colored yellow, blue, and magenta, respectively. Green spheres indicate amino acid variations identified across nine
strains of Brachyspira pilosicoli. Proteins with more than 40 variations are shown in green ribbon representation.
3.2.2.2. BP951000_RS03215 further identified a TonB-dependent receptor (TBDR)
BP951000_RS03215 is annotated as a TonB-dependent from a member of the phylum Bacteroidetes as the closest
siderophore receptor in B. pilosicoli. SignalP predicted match, supporting a role in nutrient uptake. Sequence-
a secretory signal peptide, while LipoP predicted a based annotation using PANNZER and eggNOG-
mapper, respectively, classified the protein as an OM
cytoplasmic localization. The AlphaFold 3 model revealed receptor and linked it to cobalamin transport activity
a 22-stranded C-terminal β-barrel and an N-terminal (Tables 2 and S3). Together, these results strongly suggest
plug domain (Figure 3B). DALI analysis showed the best that BP951000_RS03215 functions as a TonB-dependent
structural alignment with E. coli K-12 BtuB (PDB ID: OM receptor involved in vitamin B transport. Sequence
12
3RGM) (Tables 2 and S3), a TonB-dependent transporter variation analysis across nine B. pilosicoli strains identified
(TBDT) with a 22-stranded β-barrel and an N-terminal 59 variations (Table S5), which were mapped onto the
plug domain that occludes the lumen. E. coli BtuB structural model (Figure 3B).
111
mediates vitamin B (cobalamin) uptake via ECLs. 111-113
12
TBDTs also transport heme, ferric-siderophores, sucrose, 3.2.2.3. BP951000_RS04405
and maltodextrin. 114,115 BP951000_RS03215 shares the BP951000_RS04405 is annotated as Toxin A in UniProt
20
same overall architecture as E. coli BtuB, and its high but as a hypothetical protein in NCBI. It contains a
Z-score (32.1) confirms strong homology. Foldseek predicted secretory signal peptide. The AlphaFold 3
Volume 2 Issue 4 (2025) 94 doi: 10.36922/MI025230050