Page 90 - MI-2-4
P. 90
Microbes & Immunity Brachyspira pilosicoli novel outer membrane proteins
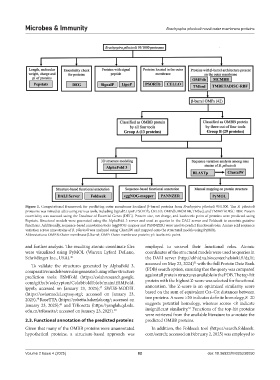
Figure 1. Computational framework for predicting outer membrane-localized β-barrel proteins from Brachyspira pilosicoli 95/1000. The B. pilosicoli
proteome was mined in silico using various tools, including SignalP, LipoP, PSORTb, CELLO, OMPdb, MCMBB, TMbed, and TMBETADISC-RBF. Protein
essentiality was assessed using the Database of Essential Genes (DEG). Protein size, net charge, and isoelectric point of proteins were predicted using
Pepstats. Structural models were generated using the AlphaFold 3 server and used as queries in the DALI server and Foldseek to annotate putative
functions. Additionally, sequence-based annotation tools (eggNOG-mapper and PANNZER) were used to predict functional roles. Amino acid sequence
variation across nine strains of B. pilosicoli was analyzed using ClustalW and mapped onto the structural models using PyMOL.
Abbreviations: OMBB: Outer membrane β-barrel; OMP: Outer membrane protein; pI: Isoelectric point.
and further analysis. The resulting atomic coordinate files employed to unravel their functional roles. Atomic
were visualized using PyMOL (Warren Lyford DeLano, coordinates of the structural models were used as queries in
Schrödinger Inc., USA). 46 the DALI server (http://ekhidna.biocenter.helsinki.fi/dali/;
51
To validate the structures generated by AlphaFold 3, accessed on May 23, 2024) with the full Protein Data Bank
comparative models were also generated using other structure (PDB) search option, ensuring that the query was compared
prediction tools: ESMFold (https://colab.research.google. against all protein structures available in the PDB. The top-hit
com/github/sokrypton/ColabFold/blob/main/ESMFold. protein with the highest Z-score was selected for functional
ipynb; accessed on January 23, 2025), SWISS-MODEL annotation. The Z-score is an optimized similarity score
47
(https://swissmodel.expasy.org/; accessed on January 23, based on the sum of equivalent Cα–Cα distances between
2025), RoseTTA (https://robetta.bakerlab.org/; accessed on two proteins. A score >20 indicates definite homology, 8–20
48.
January 23, 2025), and TrRosetta (https://yanglab.qd.sdu. suggests potential homology, whereas scores <8 indicate
49
52
edu.cn/trRosetta/; accessed on January 23, 2025). 50 insignificant similarity. Functions of the top-hit proteins
were retrieved from the available literature to annotate the
2.3. Functional annotation of the predicted proteins predicted OMBB proteins.
Given that many of the OMBB proteins were unannotated In addition, the Foldseek tool (https://search.foldseek.
hypothetical proteins, a structure-based approach was com/search; accessed on February 2, 2025) was employed to
Volume 2 Issue 4 (2025) 82 doi: 10.36922/MI025230050