Page 13 - TD-3-3
P. 13
Tumor Discovery Immune and epigenetic therapies for TNBC
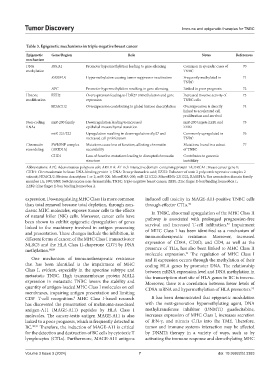
Table 3. Epigenetic mechanisms in triple‑negative breast cancer
Epigenetic Gene/Region Role Notes References
mechanism
DNA BRCA1 Promoter hypermethylation leading to gene silencing Common in sporadic cases of 70
methylation TNBC
RASSF1A Hypermethylation causing tumor suppressor inactivation Frequently methylated in 71
TNBC
APC Promoter hypermethylation resulting in gene silencing Linked to poor prognosis 72
Histone EZH2 Overexpression leading to H3K27 trimethylation and gene Increased invasive activity of 73
modification repression TNBC cells
HDAC1/2 Overexpression contributing to global histone deacetylation Overexpression is directly 74
linked to accelerated cell
proliferation and survival
Non-coding miR-200 family Downregulation leading to increased miR-200 targets ZEB1 and 75
RNAs epithelial-mesenchymal transition ZEB2
miR-221/222 Upregulation resulting in downregulation of p27 and Commonly upregulated in 76
increased cell proliferation TNBC
Chromatin SWI/SNF complex Mutations cause loss of function, affecting chromatin Mutations found in a subset 77
remodeling (ARID1A) accessibility of TNBC
CHD1 Loss of function mutations leading to disrupted chromatin Contributes to genomic
structure instability
Abbreviations: APC: Adenomatous polyposis coli; ARID1A: AT-rich interactive domain-containing protein 1A; BRCA1: Breast cancer gene 1;
CHD1: Chromodomain helicase DNA-binding protein 1; DNA: Deoxyribonucleic acid; EZH2: Enhancer of zeste 2 polycomb repressive complex 2
subunit; HDAC1/2: Histone deacetylase 1 or 2; miR-200: MicroRNA-200; miR-221/222: MicroRNA-221/222; RASSF1A: Ras association domain family
member 1A; SWI/SNF: Switch/sucrose non-fermentable; TNBC: triple-negative breast cancer; ZEB1: Zinc finger E-box binding homeobox 1;
ZEB2: Zinc finger E-box binding homeobox 2.
expression. Downregulating MHC Class I is more common induced cell toxicity in MAGE-A11-positive TNBC cells
than total removal because total depletion, through non- through effector CTLs. 92
classic MHC molecules, exposes tumor cells to the effects In TNBC, abnormal upregulation of the MHC Class II
of natural killer (NK) cells. Moreover, cancer cells have pathway is associated with prolonged progression-free
been shown to exhibit epigenetic dysregulation of genes survival and increased T-cell infiltration. Impairment
93
linked to the machinery involved in antigen processing of MHC Class I has been identified as a mechanism of
and presentation. These changes include the inhibition, in
different forms of cancer, of the MHC Class I transactivator immunotherapeutic resistance. Moreover, increased
NLRC5 and the HLA Class II-chaperone CD74 by DNA expression of CD8A, CD3D, and CD4, as well as the
methylation. 88,89 presence of TILs, has also been linked to MHC Class II
molecule expression. The regulation of MHC Class I
94
One mechanism of immunotherapeutic resistance and II expression occurs through the methylation of their
that has been identified is the impairment of MHC coding HLA genes by promoter DNA. The relationship
Class I, evident, especially in the apocrine subtype and between mRNA expression level and DNA methylation in
metastatic TNBC. High transmembrane protein MAL2 the transcription start site of HLA genes in BC is inverse.
expression in metastatic TNBC lowers the stability and Moreover, there is a correlation between lower levels of
quantity of antigen-loaded MHC Class I molecules on cell CD8A mRNA and hypermethylation of HLA promoters. 95
membranes, impairing antigen presentation and limiting
CD8 T-cell recognition. MHC Class I-based research It has been demonstrated that epigenetic modulation
9
+
has discovered the presentation of melanoma-associated with the next-generation hypomethylating agent, DNA
antigen-A11 (MAGE-A11) peptides by HLA Class I methyltransferase inhibitor (DNMTi) guadecitabine,
molecules. The cancer-testis antigen MAGE-A11 is also increases expression of MHC Class I, increases secretion
linked to a poor prognosis, which is frequently detected in of IFN-γ, and attracts CTLs into the TME. Therefore,
BC. 90,91 Therefore, the induction of MAGE-A11 is critical tumor and immune systems interaction may be affected
for the detection and destruction of BC cells by cytotoxic T by DNMTi therapy in a variety of ways, such as by
lymphocytes (CTLs). Furthermore, MAGE-A11 antigens activating the immune response and demethylating MHC
Volume 3 Issue 3 (2024) 7 doi: 10.36922/td.3383