Page 26 - EJMO-9-1
P. 26
Eurasian Journal of Medicine and
Oncology
BRCA VUS in breast cancer in MENA region
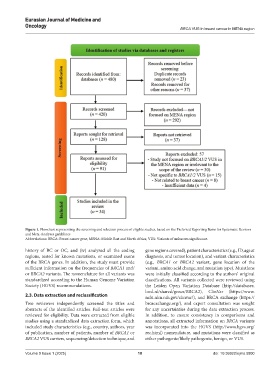
Figure 1. Flowchart representing the screening and selection process of eligible studies, based on the Preferred Reporting Items for Systematic Reviews
and Meta-Analyses guidelines
Abbreviations: BRCA: Breast cancer gene; MENA: Middle East and North Africa; VUS: Variants of unknown significance.
history of BC or OC, and (iv) analyzed all the coding gene regions covered), patient characteristics (e.g., ID, age at
regions, tested for known mutations, or examined exons diagnosis, and tumor location), and variant characteristics
of the BRCA genes. In addition, the study must provide (e.g., BRCA1 or BRCA2 variant, gene location of the
sufficient information on the frequencies of BRCA1 and/ variant, amino acid change, and mutation type). Mutations
or BRCA2 variants. The nomenclature for all variants was were initially classified according to the authors’ original
standardized according to the Human Genome Variation classifications. All variants collected were reviewed using
Society (HGVS) recommendations. the Leiden Open Variation Database (http://databases.
lovd.nl/shared/genes/BRCA2), ClinVar (https://www.
2.3. Data extraction and reclassification ncbi.nlm.nih.gov/clinvar/), and BRCA exchange (https://
Two reviewers independently screened the titles and brcaexchange.org/), and expert consultation was sought
abstracts of the identified articles. Full-text articles were for any uncertainties during the data extraction process.
reviewed for eligibility. Data were extracted from eligible In addition, to ensure consistency in comparisons and
studies using a standardized data extraction form, which annotations, all extracted information on BRCA variants
included study characteristics (e.g., country, authors, year was incorporated into the HGVS (http://www.hgvs.org/
of publication, number of patients, number of BRCA1 or rec.html) nomenclature, and mutations were classified as
BRCA2 VUS carriers, sequencing/detection technique, and either pathogenic/likely pathogenic, benign, or VUS.
Volume 9 Issue 1 (2025) 18 doi: 10.36922/ejmo.5800