Page 135 - EJMO-9-3
P. 135
Eurasian Journal of
Medicine and Oncology KRAS TP53 cholangiocarcinoma
showing a significant overall effect (Z=5.09) and a significant alterations. 11,16 The KRAS gene belongs to the RAS gene
survival difference between the mutant and wild-type groups family, and through regulation of the RAF-MEK-ERK and
(p<0.0001). A subsequent sensitivity analysis (Figure 5) PI3K-AKT-mTOR signaling pathways, it controls cellular
showed that removing individual studies did not result in growth and metabolism. Mutations in KRAS often lead to
statistically significant differences between studies, suggesting overactivation of downstream signaling pathways, thereby
that the results were robust. After addressing publication promoting tumor cell proliferation. On the other hand,
17
bias, Begg’s test and Egger’s test yielded p=0.806 and 0.570, mutations in TP53 disrupt its ability to bind to its target
respectively, indicating no publication bias (p>0.05). DNA sequence, rendering its tumor-suppressive function
ineffective. 18
4. Discussion
Current studies suggest that cancer patients with
In the field of genomic research on cholangiocarcinoma, KRAS or TP53 mutations have poorer survival outcomes
several studies have reported that KRAS and TP53 compared to those without these mutations. 19,20 In a
mutations are more common compared to other genetic meta-analysis involving stage II and III colon cancer
patients, KRAS mutations were found to be associated with
shortened DFS. Another meta-analysis on the prognostic
21
role of TP53 mutations in prostate cancer demonstrated
that patients with TP53 mutations had approximately 13%,
20%, and 16% lower OS at 1, 3, and 5 years, respectively,
compared to patients without the mutation. TP53
mutations were also associated with increased risk of death
and accelerated disease progression. 22
In our study, the results indicated that
cholangiocarcinoma patients with KRAS and TP53
mutations had a poorer prognosis compared to those
without mutations. This meta-analysis summarized
12 relevant studies, revealing that patients with KRAS
mutations had worse prognoses compared to those
without the mutation (HR=7.26; 95% CI: 6.10 – 9.81;
p<0.05). Statistically, the survival differences between the
KRAS mutant and wild-type groups were highly significant
(Z=8.39; p<0.0001). This suggests that KRAS mutations,
as an important genetic biomarker, may play a key role
in the prognosis assessment of cholangiocarcinoma. No
significant heterogeneity was found in the KRAS studies
(fixed-effects model; I =48%), supporting the use of a fixed-
2
Figure 1. Flow chart of the study design effects model for further analysis. However, Egger’s test
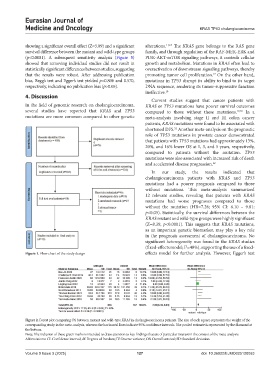
Figure 2. Forest plot comparing OS between mutant and wild-type KRAS in cholangiocarcinoma patients. The size of each square represents the weight of the
corresponding study in the meta-analysis, whereas the horizontal lines indicate 95% confidence intervals. The pooled estimate is represented by the diamond at
the bottom.
Note: The inclusion of these green marks is intended to draw attention to key findings that are of particular interest in the context of the meta-analysis.
Abbreviations: CI: Confidence interval; df: Degrees of freedom; IV: Inverse variance; OS: Overall survival; SD: Standard deviation.
Volume 9 Issue 3 (2025) 127 doi: 10.36922/EJMO025120063