Page 136 - EJMO-9-3
P. 136
Eurasian Journal of
Medicine and Oncology KRAS TP53 cholangiocarcinoma
(p<0.05) suggested potential publication bias, indicating showed that excluding individual studies did not lead to
that some negative or small sample studies may not have statistically significant differences, indicating that our
been included in the analysis. Therefore, we applied the conclusions were robust. Further testing for publication
trim-and-fill method to correct for publication bias. After bias using Begg’s test (p=0.806) and Egger’s test (p=0.570)
the correction, we found that one study was missing from showed no significant publication bias, further validating
the original analysis; adding it increased the number of the reliability of our results.
studies from 10 to 11, further confirming the significant In this study, we focused on analyzing the relationship
relationship between KRAS mutations and survival between KRAS mutations, TP53 mutations, and survival
differences (effect estimate=12.84 units, p<0.0001). This prognosis in cholangiocarcinoma patients. Although
corrected result further strengthened our conclusions.
mutations in KRAS or TP53 alone are associated with
In the analysis of TP53 gene mutations, we found poorer survival outcomes, increasing evidence suggests
that patients with TP53 mutations had significantly that the co-occurrence of KRAS and TP53 mutations
poorer prognoses compared to those without mutations may have a more pronounced effect on patient prognosis.
(HR=20.03; 95% CI: 6.79 – 33.28; p<0.05). The adverse Previous studies have indicated that, in various cancer
prognostic effect of TP53 mutations in cholangiocarcinoma types, patients with co-mutations of KRAS and TP53 often
was further statistically supported (Z=5.09; p<0.00001). exhibit more aggressive tumor behavior, poorer treatment
We observed some heterogeneity in the TP53 studies response, and shorter survival, especially in malignant
(I =59%; p=0.04), and therefore, a random-effects model cancers such as pancreatic and colorectal cancer. 23,24
2
was used for the analysis. Nevertheless, sensitivity analysis This could be due to the dual mutations, which not only
exacerbate tumor proliferation but also impair cellular self-
repair mechanisms.
While individual KRAS or TP53 mutations have been
widely studied in cholangiocarcinoma, systematic and
large-scale investigations into the biological and clinical
impact of their co-mutation remain limited. Among the
studies included in our meta-analysis, one reported a
KRAS and TP53 co-mutation rate of 16.7% in patients with
ICC. However, due to its limited sample size, the study
25
did not compare survival outcomes between patients with
co-mutations and those with only a single mutation. To
address this gap, future studies should further explore the
specific role of KRAS and TP53 co-mutations in survival
prognosis, treatment response, and drug resistance in
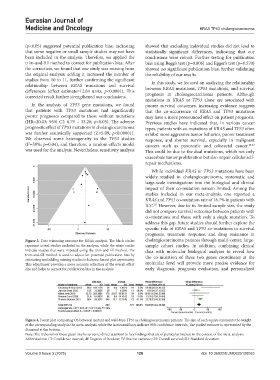
Figure 3. Data trimming outcome for KRAS analysis. The black circles cholangiocarcinoma patients through multi-center, large-
represent actual studies included in the analysis, while the white circles sample cohort studies. In addition, combining clinical
indicate studies that were imputed using the trim-and-fill method. The data with molecular biological analyses to reveal how
trim-and-fill method is used to adjust for potential publication bias by the co-mutation of these two genes coordinates at the
estimating and adding missing studies to balance funnel plot asymmetry.
This adjustment provides a more accurate reflection of the overall effect molecular level will provide more precise evidence for
size and helps to correct for publication bias in the analysis. early diagnosis, prognosis evaluation, and personalized
Figure 4. Forest plot comparing OS between mutant and wild-type TP53 in cholangiocarcinoma patients. The size of each square represents the weight
of the corresponding study in the meta-analysis, while the horizontal lines indicate 95% confidence intervals. The pooled estimate is represented by the
diamond at the bottom.
Note: The inclusion of these green marks serves to direct attention to key findings that are of particular interest in the context of the meta-analysis.
Abbreviations: CI: Confidence interval; df: Degrees of freedom; IV: Inverse variance; OS: Overall survival; SD: Standard deviation.
Volume 9 Issue 3 (2025) 128 doi: 10.36922/EJMO025120063