Page 89 - EJMO-9-3
P. 89
Eurasian Journal of
Medicine and Oncology Mapping breast cancer PPI networks for targets
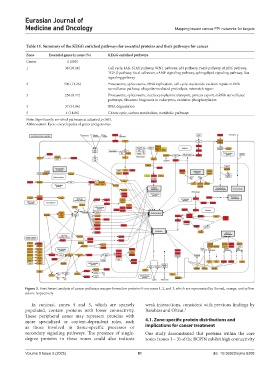
Table 11. Summary of the KEGG enriched pathways for essential proteins and their pathways for cancer
Zone Essential genes in zone (%) KEGG enriched pathways
Center 1 (100)
1 38 (10.16) Cell cycle, JAK-STAT pathway, WNT pathway, p53 pathway, FoxO pathway, MAPK pathway,
TGF-β pathway, focal adhesion, cAMP signaling pathway, sphingolipid signaling pathway, Ras
signaling pathway
2 506 (11.26) Proteasome, spliceosome, DNA replication, cell cycle, nucleotide excision repair, mRNA
surveillance pathway, ubiquitin-mediated proteolysis, mismatch repair
3 256 (8.77) Proteasome, spliceosome, nucleocytoplasmic transport, protein export, mRNA surveillance
pathways, ribosome biogenesis in eukaryotes, oxidative phosphorylation
4 37 (14.86) RNA degradation
5 4 (14.86) Citrate cycle, carbon metabolism, metabolic pathways
Note: Significantly enriched pathways at adjusted p<0.05.
Abbreviation: Kyoto encyclopedia of genes and genomes.
Figure 3. Enrichment analysis of cancer pathways was performed on proteins from zones 1, 2, and 3, which are represented by the red, orange, and yellow
colors, respectively
In contrast, zones 4 and 5, which are sparsely weak interactions, consistent with previous findings by
populated, contain proteins with lower connectivity. Barabási and Oltvai. 3
These peripheral zones may represent proteins with
more specialized or context-dependent roles, such 4.1. Zone-specific protein distributions and
as those involved in tissue-specific processes or implications for cancer treatment
secondary signaling pathways. The presence of single- Our study demonstrated that proteins within the core
degree proteins in these zones could also indicate zones (zones 1 – 3) of the BCPIN exhibit high connectivity
Volume 9 Issue 3 (2025) 81 doi: 10.36922/ejmo.8208