Page 9 - GPD-2-1
P. 9
Gene & Protein in Disease AI-based drug repositioning
and drugs in heterogeneous networks more easily and was used to extract protein features from five ion channel
efficiently. Chen et al. invented a heterogeneous network target proteins screened from the SARS-CoV-2 whole-
supporting different relationships, which include drugs and genome sequence of the NCBI database. The extracted
proteins linked by known drug-target interaction, chemical drug features were then connected with the target feature
similarity between drugs, and sequence similarity between information using graph convolutional network (GCN)
proteins to mine potential drug-disease associations . In and attention mechanism. The drug-target affinity is
[10]
2018, Olayan RS used a nonlinear fusion approach to fuse outputted after two layers of fully connected operation,
drug-protein features from different similarity networks and finally the drug-target affinity model is obtained. One
and pathway-based features in these networks together . of the hybrid graphs network-based drug-target affinity
[11]
Peng et al. completed random wandering with restarts prediction model framework is shown in Figure 2.
using a similarity-based heterogeneous network model,
and based on that, a denoising autoencoder was used to 3. Feature-based approach
implement a convolutional neural network predictive The feature extraction method uses a new feature space of
classifier and learn the basic features . Ji et al. used a lower dimensionality to map the original feature projection,
[12]
heterogeneous network combining a large amount of while the new features are usually a combination of the
functional information and a large amount of information original features, with the aim of finding more meaningful
about the structure of the network nodes to perfectly information. The common feature extraction techniques
solve the problem of excessive feature generation by are principal component analysis and singular value
[17]
the nodes in the heterogeneous network to calculate the decomposition . The purpose of the selection method
final feature vector . Lu et al. investigated a drug-target is to select small portions of features from the complete
[13]
interaction prediction method based on multisource data set of input features based on some design criteria to be
fusion and network structure perturbation . He et al. used as input to the model. In the process of predicting
[14]
invented and disclosed a computational drug relocation drug sensitivity, a priori biological knowledge is usually
method based on memory networks and attention . incorporated into the feature fraction.
[15]
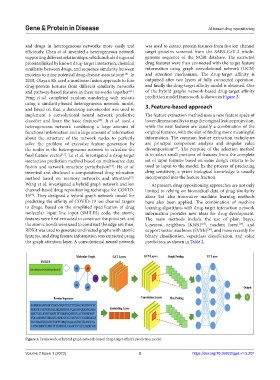
Wang et al. investigated a hybrid graph network and ion At present, drug repositioning approaches are not only
channel-based drug repositioning technique for COVID- limited to relying on biomedical data of drug similarity
19 . They designed a hybrid graph network model for alone but also innovative machine learning methods
[16]
predicting the affinity of COVID-19 ion channel targets have also been applied. The combination of machine
to drugs. Based on the simplified specification of drug learning algorithms with drug-target interaction network
molecular input line input (SMILES) code, the atomic information provides new ideas for drug development.
features were first extracted to construct the point set, and The main methods include the use of plain Bayes,
the atomic bonds were used to construct the edge set; then, k-nearest neighbors (KNN) , random forest , and
[18]
[19]
RDKit was used to generate undirected graphs with atomic support vector machines (SVMs) , and more recently for
[20]
features, and drug feature information was extracted using binary classification, superclass classification, and value
the graph attention layer. A convolutional neural network prediction, as shown in Table 2.
Figure 2. Framework of hybrid graph network-based drug-target affinity prediction model.
Volume 2 Issue 1 (2023) 3 https://doi.org/10.36922/gpd.v1i3.201