Page 11 - GPD-2-1
P. 11
Gene & Protein in Disease AI-based drug repositioning
A
B
C
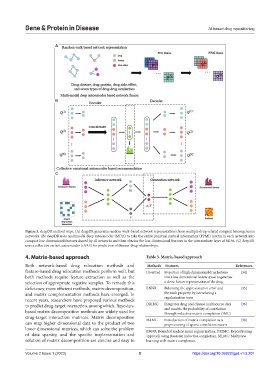
Figure 3. deepDR method steps. (A) deepDR generates random walk-based network representations from multiple drug-related complex heterogeneous
networks. (B) deepDR uses multimodal deep autoencoder (MDA) to take the entire punctual mutual information (PPMI) matrix in each network into
compact low-dimensional features shared by all networks and then obtains the low-dimensional features in the intermediate layer of MDA. (C) deepDR
uses a collective variant autoencoder (cVAE) for prediction of disease-drug relationships.
4. Matrix-based approach Table 3. Matrix‑based approach
Both network-based drug relocation methods and Methods Features References
feature-based drug relocation methods perform well, but DivePred Projection of high-dimensional drug features [34]
both methods require feature extraction as well as the into a low-dimensional feature space to generate
selection of appropriate negative samples. To remedy this a dense feature representation of the drug
deficiency, more efficient methods, matrix decomposition, BNNR Balancing the approximation error and [35]
and matrix complementation methods have emerged. In the rank property by introducing a
recent years, researchers have proposed various methods regularization term
to predict drug-target interaction, among which, Bayesian- DRIMC Integrates drug and disease multisource data [36]
based matrix decomposition methods are widely used for and models the probability of correlation
through inductive matrix completion (IMC)
drug-target interaction matrices. Matrix decomposition MLMC Introduction of matrix completion as a [38]
can map higher dimensional data to the product of two preprocessing of sparse correlation matrix
lower dimensional matrices, which can solve the problem BNNR: Bounded nuclear norm regularization, DRIMC: Repositioning
of data sparsity, and the specific implementation and approach using Bayesian inductive completion, MLMC: Multiview
solution of matrix decomposition are concise and easy to learning with matrix completion.
Volume 2 Issue 1 (2023) 5 https://doi.org/10.36922/gpd.v1i3.201