Page 77 - GPD-2-3
P. 77
Gene & Protein in Disease Hotspots in the FOXO4: p53 interaction
Table 1. Amino acid sequences and theoretical and experimental m/z (monoisotopic) of peptides tested in this work
Entry Sequence Theoretical (MW) Experimental (MW)
FOXO4-DRI NH -tlrkepaseiaqsileaysqngwanrr-OH 3087.54 3087.546
2
FOXO4-DRI_short NH -leaysqngw-OH 1066.44 1066.461
2
FOXO4 NH -RRNAWGNQSYAELISQAIESAPEKRLT-OH 3087.54 3087.546
2
FOXO4_short NH -WGNQSYAEL-OH 1066.44 1066.461
2
Notes: All peptides are free at N-terminus and COOH at C-terminus. D-amino acids are shown in lowercase to distinguish them from L-amino acids
(uppercase).
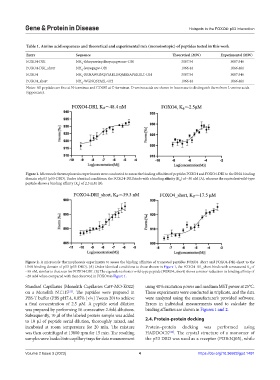
Figure 1. Microscale thermophoresis experiments were conducted to assess the binding affinities of peptides FOXO4 and FOXO4-DRI to the DNA binding
domain of p53 (p53-DBD). Under identical conditions, the FOXO4-DRI binds with a binding affinity (K ) of ~50 nM (A), whereas the equivalent wild-type
d
peptide shows a binding affinity (K ) of 2.5 mM (B).
d
Figure 2. A microscale thermophoresis experiments to assess the binding affinities of truncated peptides FOXO4_short and FOXO4-DRI-short to the
DNA binding domain of p53 (p53-DBD). (A) Under identical conditions to those shown in Figure 1, the FOXO4-DR_short binds with a measured K of
d
~50 nM, similar to that seen for FOXO4-DRI. (B) The equivalent shorter wild-type peptide (FOXO4_short) shows a minor reduction in binding affinity of
~20 mM when compared with that observed in FOXO4 in Figure 1.
Standard Capillaries (Monolith Capillaries Cat#-MO-K022) using 40% excitation power and medium MST power at 25°C.
on a Monolith NT.115 . The peptides were prepared in These experiments were conducted in triplicate, and the data
[21]
PBS-T buffer (PBS pH7.4, 0.05% [v/v] Tween 20) to achieve were analyzed using the manufacturer’s provided software.
a final concentration of 2.5 μM. A peptide serial dilution Errors in individual measurements used to calculate the
was prepared by performing 16 consecutive 2-fold dilutions. binding affinities are shown in Figures 1 and 2.
Subsequently, 10 μl of the labeled protein sample was added
to 10 μl of peptide serial dilution, thoroughly mixed, and 2.4. Protein-protein docking
incubated at room temperature for 20 min. The mixture Protein–protein docking was performed using
was then centrifuged at 13000 rpm for 15 min. The resulting HADDOCK [22] . The crystal structure of a monomer of
samples were loaded into capillary trays for data measurement the p53 DBD was used as a receptor (PDB:3Q05), while
Volume 2 Issue 3 (2023) 4 https://doi.org/10.36922/gpd.1491