Page 78 - GPD-2-3
P. 78
Gene & Protein in Disease Hotspots in the FOXO4: p53 interaction
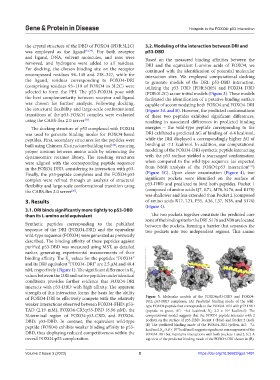
the crystal structure of the DBD of FOXO4 (PDB:3L2C) 3.2. Modeling of the interaction between DRI and
was employed as the ligand [23,24] . For both receptor p53-DBD
and ligand, DNA, solvent molecules, and ions were Based on the measured binding affinities between the
removed, and hydrogens were added to all residues. DRI and the equivalent L-amino acids of FOXO4, we
For docking, the chosen binding site on the receptor continued with the identification of potential molecular
encompassed residues 94–148 and 238–327, while for interaction sites. We employed computational docking
the ligand, residues corresponding to FOXO4-DRI to generate models of the DRI: p53-DBD interaction,
(comprising residues 93–119 of FOXO4 in 3L2C) were utilizing the p53 DBD (PDB:3Q05) and FOXO4 DBD
selected to form the PPI. The p53-FOXO4 pose with (PDB:3L2C) as our initial models (Figure 3). These models
the best complementarity between receptor and ligand facilitated the identification of a putative binding surface
was chosen for further analysis. Following docking, capable of accommodating both FOXO4 and FOXO4-DRI
the structural flexibility and large-scale conformational (Figure 3A and B). However, the predicted conformations
transitions of the p53-FOXO4 complex were evaluated of these two peptides exhibited significant differences,
using the CABS-flex 2.0 server [25] . resulting in associated differences in predicted binding
The docking structure of p53 complexed with FOXO4 energies – the wild-type peptide corresponding to the
was used to generate binding modes for FOXO4-based DRI exhibited a predicted ΔG of binding of -6.4 kcal/mol,
peptides. First, secondary structures for the peptides were while the DRI displayed a correspondingly higher ΔG of
built using Chimera X’s structure building tool , ensuring binding at -7.1 kcal/mol. In addition, our computational
[26]
proper torsions between amino acids by referencing the modeling of the FOXO4-DRI synthetic peptide interacting
dynameomics rotamer library. The resulting structures with the p53 surface yielded a rearranged conformation
were aligned with the corresponding peptide sequence when compared to the wild-type sequence (as expected
[21]
in the FOXO4 DBD, considering its interaction with p53. from NMR analysis of the FOXO4:p53 interaction )
Finally, the p53-peptide complexes and the FOXO4:p53 (Figure 3C). Upon closer examination (Figure 4), two
complex were refined through an analysis of structural significant pockets were identified on the surface of
flexibility and large-scale conformational transition using p53-DBD and predicted to bind both peptides. Pocket 1
[25]
the CABS-flex 2.0 server . (composed of amino acids Q7, K71, M76, S176, and E178)
was shallower and less extended than Pocket 2 (composed
3. Results of amino acids R17, L21, P35, A36, L37, N38, and S176)
(Figure 4).
3.1. DRI binds significantly more tightly to p53-DBD
than its L-amino acid equivalent The two pockets together constitute the predicted core
zone of the binding site for the DRI. S176 and N38 are located
Synthetic peptides corresponding to the published between the pockets, forming a barrier that separates the
sequence of the DRI (FOXO4-DRI) and the equivalent two pockets into two independent regions. This amino
wild-type sequence (FOXO4) were generated as previously
described. The binding affinity of these peptides against
purified p53-DBD was measured using MST, as detailed
earlier, generating experimental measurements of their
binding affinity. The K values for the peptides “FOXO4”
d
and its DRI equivalent “FOXO4-DRI” are 2.5 μM and 48.4
nM, respectively (Figure 1). The significant difference in K
d
values between the DRI and native peptides under identical
conditions provides further evidence that FOXO4-DRI
interacts with p53-DBD with high affinity. The apparent
strength of this interaction forms the basis for the ability
of FOXO4-DRI to effectively compete with the relatively Figure 3. Molecular models of the FOXO4:p53-DBD and FOXO4-
DRI: p53-DBD complexes. (A) Predicted binding mode of the wild-
weaker interactions observed between FOXO4-FHD: p53- type FOXO4 peptide that corresponds to the FOXO4-DRI with p53-DBD
TAD (2.19 mM), FOXO4-CR3:p53-DBD (6.96 μM), the (peptide in green, ΔG: −6.4 kcal/mol, K : 2.2 × 10 kcal/mol). The
-5
d
N-terminal region of FOXO4:p53-CRD, and FOXO4- computational model suggests that the FOXO4 peptide interacts with 2
DBD: p53-DBD. In contrast, the equivalent wild-type pockets on the surface of p53-DBD: Pocket 1 (blue) and Pocket 2 (red).
peptide (FOX04) exhibits weaker binding affinity to p53- (B) The predicted binding mode of the FOXO4-DRI (yellow, ΔG: −7.1
kcal/mol, K : 5.8 × 10 kcal/mol) suggests significant rearrangement of the
-6
DBD, thus displaying reduced competitiveness within the FOXO4-DRI but maintains interactions with both pockets 1 and 2. (C) A
d
overall FOXO4:p53 complexation. top view of the predicted binding mode of the FOXO4-DRI shown in (B).
Volume 2 Issue 3 (2023) 5 https://doi.org/10.36922/gpd.1491