Page 119 - GPD-4-2
P. 119
Gene & Protein in Disease Pediatric glioma circadian clock genes
A B
D
C
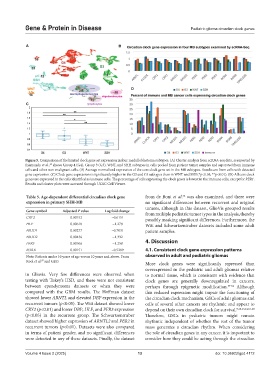
Figure 5. Comparison of the limited clock gene set expression in four medulloblastoma subtypes. (A) Cluster analysis from scRNA-seq data, as reported by
Riemondy et al., shows Group 4 (G4), Group 3 (G3), WNT, and SHH subtypes in cells pooled from patient tumor samples and separated from immune
49
cells and other non-malignant cells. (B) Average normalized expression of the core clock gene set in the MB subtypes. Results are from cells with detected
gene expression. (C) Clock gene expression is significantly higher in the G3 and G4 subtypes than in WNT and SSH (*p<0.05, **p<0.01). (D) All core clock
genes are expressed in the cells identified as immune cells. The percentage of cells expressing the clock genes is lowest in the immune cells, except for PER1.
Results and cluster plots were accessed through UCSC Cell Viewer.
54
Table 5. Age‑dependent differential circadian clock gene from de Bont et al. was also examined, and there were
expression in primary SHH‑MB no significant differences between recurrent and original
tumors, although in this dataset, GlioVis grouped results
Gene symbol Adjusted P value Log fold change from multiple pediatric tumor types in the analysis, thereby
CRY2 0.00152 −0.6151 possibly masking significant differences. Furthermore, the
HLF 0.00618 −1.278 Witt and Schwartzentruber datasets included some adult
NR1D1 0.00277 −0.7031 patient samples.
NR1D2 0.00436 −1.332
PER3 0.00106 −1.258 4. Discussion
RORA 0.00151 −0.5289 4.1. Consistent clock gene expression patterns
Note: Patients under 10 years of age versus 10 years and above. From observed in adult and pediatric gliomas
Kool et al. and GEO.
57
More clock genes were significantly repressed than
overexpressed in the pediatric and adult gliomas relative
in Gliovis. Very few differences were observed when to normal tissue, which is consistent with evidence that
testing with Tukey’s HSD, and these were not consistent clock genes are generally downregulated in cancers,
between ependymoma datasets or when they were perhaps through epigenetic modification. 59-62 Although
compared with the GBM results. The Hoffman dataset this reduced expression might impair the functioning of
showed lower ARNTL and elevated DBP expression in the the circadian clock mechanism, GSCs of adult gliomas and
recurrent tumors (p<0.05). The Witt dataset showed lower cells of several other cancers are rhythmic and appear to
CRY2 (p<0.01) and lower DBP, HLF, and PER3 expression depend on their own circadian clock for survival. 9,10,13,14,63-68
(p<0.05) in the recurrent group. The Schwartzentruber Therefore, GSCs in pediatric tumors might remain
dataset showed higher expression of ARNTL2 and PER2 in rhythmic independent of whether the rest of the tumor
recurrent tumors (p<0.01). Datasets were also compared mass generates a circadian rhythm. When considering
in terms of patient gender, and no significant differences the role of circadian genes in any cancer, it is important to
were detected in any of these datasets. Finally, the dataset consider how they could be acting through the circadian
Volume 4 Issue 2 (2025) 13 doi: 10.36922/gpd.4112