Page 118 - GPD-4-2
P. 118
Gene & Protein in Disease Pediatric glioma circadian clock genes
Table 4. (Continued)
Cavalli, 2017 Robinson, 2012 Northcott, 2011
G3 G3 C
G4 G4 D
RORB RORB RORB
WNT SHH G3 WNT SHH G3 WNT SHH C
SHH SHH SHH
G3 G3 C
G4 G4 D
TEF TEF TEF
WNT SHH G3 WNT SHH G3 WNT SHH C
SHH SHH SHH
G3 G3 C
G4 G4 D
TIMELESS TIMELESS TIMELESS
WNT SHH G3 WNT SHH G3 WNT SHH C
SHH SHH SHH
G3 G3 C
G4 G4 D
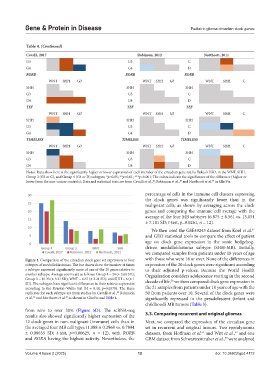
Notes: Data show here is the significantly higher or lower expression of each member of the circadian gene set, by Tukey’s HSD, in the WNT, SHH,
Group 3 (G3 or C), and Group 4 (G4 or D) subtypes; *p<0.05; **p<0.01; ***p<0.001; The colors indicate the significance of the difference (higher or
lower from the non-cancer controls); Data and statistical tests are from Cavalli et al., ; Robinson et al., and Northcott et al. in GlioVis.
46
47
45
percentage of cells in the immune cell clusters expressing
the clock genes was significantly lower than in the
malignant cells, as shown by averaging across the clock
genes and comparing the immune cell average with the
average of the four MB subtypes (6.875 ± 6.561 vs. 13.054
± 7.181 SD; t-test, p=0.0261, n = 12).
We then used the GSE49243 dataset from Kool et al.
57
and GEO statistical tools to compare the effect of patient
age on clock gene expression in the sonic hedgehog-
driven medulloblastoma subtype (SHH-MB). Initially,
we compared samples from patients under 18 years of age
Figure 4. Comparison of the circadian clock gene set expression in four with those who were 18 or over. None of the differences in
subtypes of medulloblastomas. The bar charts show the number of times expression of the 20 clock genes were significant according
a subtype expressed significantly more of one of the 20 genes relative to to their adjusted p-values. Because the World Health
another subtype. Average counts are as follows: Group 4 = 24 (± 3.60 SD); Organization considers adolescence starting in the second
Group 3 = 10.33 (± 3.21 SD); WNT = 6.67 (± 3.21 SD); and SHH = 6 (± 1 58
SD). The subtypes have significant differences in their relative expression decade of life, we then compared clock gene expression in
according to the Kruskal–Wallis test (H = 8.44, p=0.0378). The three the 21 samples from patients under 10 years of age with the
replicates for each subtype are from studies by Cavalli et al., Robinson 50 from patients over 10. Several of the clock genes were
45
et al., and Northcott et al. as shown in GlioVis and Table 4. significantly repressed in the preadolescent (infant and
47
46
childhood) MB tumors (Table 5).
from zero to over 30% (Figure 5D). The scRNA-seq
results also showed significantly higher expression of the 3.3. Comparing recurrent and original gliomas
12 clock genes in non-malignant (immune) cells than in Next, we compared the expression of the circadian gene
the averaged four MB cell types (1.088 ± 0.2968 vs. 0.7894 set in recurrent and original tumors. Two ependymoma
± 0.08655 SD; t-test, p=0.00625, n = 12), with RORB datasets, from Hoffman et al. and Witt et al., and one
52
51
and RORA having the highest activity. Nevertheless, the GBM dataset from Schwartzentruber et al., were analyzed
53
Volume 4 Issue 2 (2025) 12 doi: 10.36922/gpd.4112