Page 99 - GPD-4-2
P. 99
Gene & Protein in Disease Significance of MXRA7 in bladder cancer
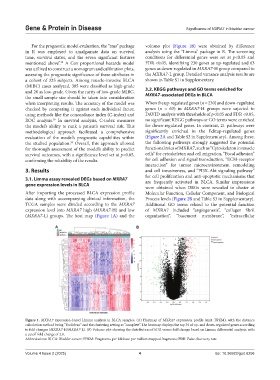
For the prognostic model evaluation, the “rms” package volcano plot (Figure 1B) were obtained by difference
in R was employed to amalgamate data on survival analysis using the “Limma” package in R. The screening
time, survival status, and the seven significant features conditions for differential genes were set at p<0.05 and
mentioned above”. A Cox proportional hazards model FDR <0.05, identifying 230 genes as up-regulated and 63
38
was utilized to construct a nomogram and calibration plots, genes as down-regulated in MXRA7-H group compared to
assessing the prognostic significance of these attributes in the MXRA7-L group. Detailed variance analysis results are
a cohort of 325 subjects. Among muscle-invasive BLCA shown in Table S1 in Supplementary.
(MIBC) cases analyzed, 305 were classified as high-grade
and 20 as low-grade. Given the rarity of low-grade MIBC, 3.2. KEGG pathways and GO terms enriched for
the small sample size should be taken into consideration MXRA7-associated DEGs in BLCA
when interpreting results. The accuracy of the model was When the up-regulated genes (n = 230) and down-regulated
checked by comparing it against each individual factor, genes (n = 63) in MXRA7-H groups were sujected to
using methods like the concordance index (C-index) and DAVID analysis with thresholds of p<0.05 and FDR <0.05,
ROC analysis. In survival analysis, C-index measures no significant KEGG pathways or GO terms were enriched
39
the model’s ability to rank a patient’s survival risk. This for down-regulated genes. In contrast, 21 pathways were
methodological approach facilitated a comprehensive significantly enriched in the Editup-regulated genes
evaluation of the model’s prognostic capabilities within (Figure 2A and Table S2 in Supplementary). Among these,
the studied population. Overall, this approach allowed the following pathways strongly suggested the potential
39
for thorough assessment of the model’s ability to predict functional roles of MXRA7, such as “Cytoskeleton in muscle
survival outcomes, with a significance level set at p<0.05, cells” for cytoskeleton and cell migration, “Focal adhesion”
confirming the reliability of the results. for cell adhesion and signal transduction, “ECM-receptor
interaction” for tumor microenvironment remodeling
3. Results and cell invasiveness, and “PI3K-Akt signaling pathway”
for cell proliferation and anti-apoptotic mechanisms that
3.1. Limma assay revealed DEGs based on MXRA7 are frequently activated in BLCA. Similar impressions
gene expression levels in BLCA
were obtained when DEGs were revealed to cluster at
After importing the processed BLCA expression profile Molecular Function, Cellular Component, and Biological
data along with accompanying clinical information, the Process levels (Figure 2B and Table S3 in Supplementary).
TCGA samples were divided according to the MXRA7 Additional GO terms related to the potential function
expression level into MXRA7 high (MXRA7-H) and low of MXRA7 included “angiogenesis”, “collagen fibril
(MXRA7-L) groups. The heat map (Figure 1A) and the organization”, “basement membrane”, “extracellular
A B
Figure 1. MXRA7 expression-based Limma analysis in BLCA samples. (A) Heatmap of MXRA7 expression profile (unit: FPKM), with the distance
calculation method being “Euclidean” and the clustering setting as “complete”. The heatmap displays the top 30 of up- and down-regulated genes according
to fold changes (MXRA7-H/MXRA7-L). (B) Volcano plot showing the distribution of FDR versus fold change based on Limma differential analysis, with
a cutoff fold change of 2.0.
Abbreviations: BLCA: Bladder cancer; FPKM: Fragments per kilobase per million mapped fragments; FDR: False discovery rate.
Volume 4 Issue 2 (2025) 4 doi: 10.36922/gpd.6256