Page 34 - GTM-2-4
P. 34
Global Translational Medicine The research advances in HPV integration
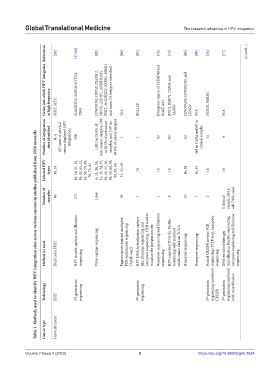
References [58] [57,94] [95] [96] [97] [73] [75] [86] [98] [76] [77] (Cont’d...)
Genes into which HPV integrates at high frequency FHIT, MYC MACROD2, MIPOLA/TTC6, TP63 LINC00392, LRP1B, DIAPH 2, PROS1, LSP1P5, ANKRD26P1, FHIT, MACROD2, ERBB2, RREB1 and more (24 hotspots identified) N/A BCL11B Intergenic region of CHMP48 and RALY‑AS1 BNC1, RSBN1, USP36, and TAOK3 LINC00290, LINC02500, and LENG9 N/A NISCH, PBRM1 N/A
Number of integration sites identified N/A (62 cases of cervical cancer displayed HPV integration) 308 1,002 in 24.8% of non-cancer samples, 588 in 38.0% of precancer samples, and 1597 in 69.0% of cancer samples 1 1 83 267 63 448 in CaSki and 60 in clinical samples 74 4
Table 1. Methods used to identify HPV integration sites across various cancers in studies published from 2020 onwards
Infected HPV types 16, 18 16, 18, 31, 33, 39, 42, 45, 52, 56, 58, 59, 68, 70, 73, 82 6, 11, 16, 18, 31, 33, 34, 35, 39, 45, 52, 56, 58, 59, 66, 68, 69, 82, etc 51, 52, 59 70 16 16 16, 58 16, 35 18 18
Number of samples 96 272 1,466 36 1 1 8 16 1 2 0 clinical sample, HeLa cell DNA used
HPV double capture and Illumina Tagmentation-assisted multiplex PCR enrichment sequencing HPV DNA hybridization capture plus Illumina sequencing and nanopore sequencing, FISH used to visualize the integration site Nanopore sequencing and Illumina HPV-enriched DNA for PacBio multi-omics data on TCGA Pooled CRISPR inverse PCR sequencing (PCIP-seq), nanopore microfluidics, PacBio sequencing, nanopore sequencing and Illumina
Method (s) used Dual-color FISH sequencing Virus capture sequencing (TaME-seq2) sequencing sequencing with matched Nanopore sequencing Nanopore sequencing sequencing DNA enriched through sequencing
Technology FISH 2 nd -generation sequencing 3 rd -generation sequencing 3 rd -generation sequencing combined CRISPR 3 rd -generation sequencing combined with microfluidics
Cancer type Cervical cancer
Volume 2 Issue 4 (2023) 8 https://doi.org/10.36922/gtm.2034