Page 148 - v11i4
P. 148
International Journal of Bioprinting AI for sustainable bioprinting
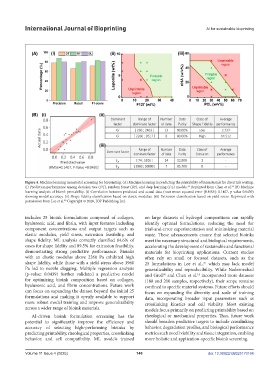
Figure 4. Machine learning in material screening for bioprinting. (A) Machine learning in predicting the printability of biomaterials for direct ink writing.
(i) Prediction performance among decision tree (DT), random forest (RF), and deep learning (DL) models. Reprinted from Chen et al. (B) Machine
14
14
learning analysis of bioink printability. (i) Correlation between predicted and actual data (root mean squared error [RMSE]: 0.1407, p-value 0.0439)
showing model accuracy. (ii) Shape fidelity classification based on elastic modulus. (iii) Extrusion classification based on yield stress. Reprinted with
permission from Lee et al. Copyright © 2020, IOP Publishing Ltd.
80
includes 25 bioink formulations composed of collagen, on large datasets of hydrogel compositions can rapidly
hyaluronic acid, and fibrin, with input features including identify optimal formulations, reducing the need for
component concentrations and output targets such as trial-and-error experimentation and minimizing material
elastic modulus, yield stress, extrusion feasibility, and waste. These advancements ensure that selected bioinks
shape fidelity. ML analysis correctly classified 84.6% of meet the necessary structural and biological requirements,
cases for shape fidelity and 89.5% for extrusion feasibility, accelerating the development of sustainable and functional
demonstrating strong predictive performance. Bioinks materials for bioprinting applications. Current studies
with an elastic modulus above 2260 Pa exhibited high often rely on small or focused datasets, such as the
shape fidelity, while those with a yield stress above 3960 25 formulations in Lee et al., which may lack model
81
Pa led to nozzle clogging. Multiple regression analysis generalizability and reproducibility. While Nadernezhad
(p-value: 0.0439) further validated a predictive model and Groll and Chen et al. incorporated more datasets
14
80
for optimizing bioink composition based on collagen, (180 and 210 samples, respectively), their scope remains
hyaluronic acid, and fibrin concentrations. Future work confined to specific material systems. Future efforts should
can focus on expanding the dataset beyond the initial 25 focus on expanding the diversity and scale of training
formulations and making it openly available to support data, incorporating broader input parameters such as
more robust model training and improve generalizability crosslinking kinetics and cell viability. Most existing
across a wider range of bioink materials. models focus primarily on predicting printability based on
AI-driven bioink formulation screening has the rheological or mechanical properties. Thus, future work
potential to significantly improve the efficiency and should broaden predictive targets to include crosslinking
accuracy of selecting high-performing bioinks by behavior, degradation profiles, and biological performance
predicting printability, rheological properties, crosslinking metrics such as cell viability and tissue integration, enabling
behavior, and cell compatibility. ML models trained more holistic and application-specific bioink screening.
Volume 11 Issue 4 (2025) 140 doi: 10.36922/IJB025170164