Page 73 - ITPS-7-3
P. 73
INNOSC Theranostics and
Pharmacological Sciences Enhancers and SEs in cancer treatment
RNAs (eRNAs), and histone-modifying enzymes including become active solely through H3K4me1 modification.
9,10
methyltransferases, histone acetyltransferase EP300, and Genes linked to the H3K27ac enhancer mark exhibit
CBP. This cooperative binding initiates and promotes higher expression levels compared to those associated with
transcription (Figure 1A). Although most enhancers are the H3K4me1 enhancer mark. Further studies described
11
2-4
located in intergenic and intronic regions of the genome, CBP/EP300-mediated H3K27 acetylation as a marker
some are located within exons. Super-enhancers (SEs) are of active enhancers, since repressing this modification
5
extensive genomic regions formed by clusters of enhancers. reduces enhancer activity, indicating that H3K27ac is
SEs exhibit a higher (several-fold) binding enrichment for causative, not just correlative, to enhancer activity. An
12
transcriptional factors than typical enhancers, spanning illustration of this concept is the discovery that EP300
more than 20 kb on average. SEs have a greater impact regulates enhancers in neuroblastoma (NB) by adding the
6
on the transcription of specific genes in comparison to H3K27ac mark to colorectal cancer-associated SEs. This
regular enhancers and have the ability to simultaneously process involves interaction with the recently identified TF
activate a significant number of promoters. These SEs TFAP2β in NB cells. Moreover, EP300 has been shown
13
are typically found in close proximity to genes crucial to disrupt the activity of epigenetic modifiers known to
for cell differentiation. Scientific findings suggest that regulate enhancers, such as histone deacetylases and non-
7,8
active enhancers are often marked by the co-occurrence of coding RNA (ncRNAs), hence promoting pulmonary
H3K4me1 and H3K27ac. However, certain enhancers can fibrosis. Intriguingly, genome-wide RNA sequencing has
14
A
B
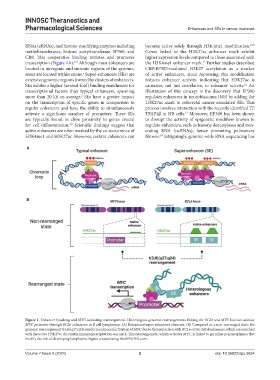
Figure 1. Enhancer hijacking and MYC-activating rearrangement. Heterologous genomic rearrangements linking the BCL6 and MYC loci can activate
MYC promoter through BCL6 enhancers in B cell lymphomas. (A) Enhancers/super-enhancers elements. (B) Compared to a non-rearranged state, the
genomic rearrangement t(3;8)(q27;q24) results in enhanced activation of MYC due to the interaction with BCL6 active distal enhancers, which are enriched
with the active H3K27ac chromatin immunoprecipitation-seq mark. This rearrangement, which activates MYC, is linked to germline polymorphisms that
modify the risk of developing lymphoma. Figure created using BioRENDER.com.
Volume 7 Issue 3 (2024) 2 doi: 10.36922/itps.3654