Page 24 - JCTR-11-5
P. 24
Journal of Clinical and
Translational Research AI and LLMs in iPSC cardiac research
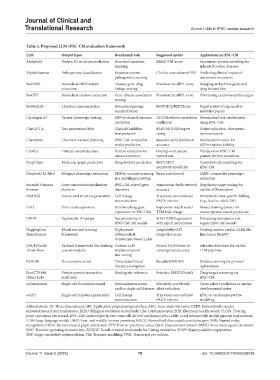
Table 5. Proposed LLM‑iPSC‑CM evaluation framework
LLM Output types Benchmark task Suggested metric Application in iPSC‑CM
AlphaFold Protein 3D structure prediction Structural mutation RMSD/TM-score Sarcomeric protein modeling for
mapping inherited cardiac diseases
AlphaMissense Pathogenicity classification Missense variant ClinVar concordance/PPV Predicting clinical impact of
pathogenicity scoring sarcomeric mutations
BioBERT Biomedical NER/relation Disease-gene-drug Precision/recall/F1 score Mapping arrhythmia genes and
extraction linkage mining drug interactions
BioGPT Biomedical relation extraction Gene-disease association Precision/recall/F1 score Prioritizing cardiomyopathy targets
mining
BioMedLM Literature summarization Biomedical passage ROUGE-L/BERTScore Rapid review of regenerative
summarization medicine papers
Cardiogen AI Variant-phenotype linking SNP-to-clinical outcome AUC/Matthews correlation Personalized risk stratification
prediction coefficient using iPSC-CM
ChatGPT-4 Text generation/Q&A Clinical Guideline BLEU/ROUGE/expert Patient education, therapeutic
interpretation rating summarization
Chemputer Chemical reaction planning iPSC-CM-compatible Reaction yield prediction Media optimization for
media prediction accuracy differentiation/stability
ClinVar Clinical variant database Variant validation for Overlap with patient Validation of iPSC-CM
disease relevance variant sets patient-derived mutations
DeepChem Molecular graph prediction Drug-toxicity prediction ROC/AUC/ Cardiotoxicity modeling via
sensitivity-specificity iPSC-CM
DeepSeek-R1/Med Bilingual phenotype extraction EHR-to-concept mapping Exact match/recall LMIC-compatible phenotype
in a multilingual setting extraction
Ensembl Genome Gene annotation/visualization iPSC-CM-related gene Annotation depth/retrieval Regulatory target mining for
Browser platform discovery accuracy cardiac differentiation
ESMFold End-to-end structure generation Cell lineage Trajectory concordance/ Maturation-state-specific folding
reconstruction PAGA metrics (e.g., fetal vs. adult CM)
GEO Omics data repository Benchmarking gene Expression match score/ Model training dataset for
expression in iPSC-CMs TPM fold-change transcriptomic-based prediction
GROK Explainable AI output Interpretability of SHAP/LIME agreement Enhancing transparency in
iPSC-CM risk models with expert annotations regenerative risk models
HuggingFace Model zoo and training Deployment Adaptability/API Hosting custom cardiac LLMs like
Transformers framework of biomedical integration score fine-tuned BioGPT
transformer-based LLMs
JAX/PyTorch/ Backend frameworks (for training Custom LLM Neural FLOPs/time to Infrastructure layer for cardiac
TensorFlow custom models) implementation/ convergence/accuracy LLM pipelines
fine-tuning
REALM Document retrieval Omics data-linked Recall@10/NDCG Evidence mining for protocol
literature navigation optimization
RoseTTAfold Protein-protein interaction Binding site inference Interface RMSD/DockQ Drug-target screening via
(Baker Lab) prediction iPSC-CM
scFoundation Single-cell foundation model Generalization across Silhouette score/batch Cross-cohort prediction in cardiac
cardiac single-cell datasets effect reduction developmental states
scGPT Single-cell trajectory generation Cell lineage Trajectory concordance/ iPSC-to-cardiomyocyte fate
reconstruction PAGA metrics modeling
Abbreviations: 3D: Three-dimensional; API: Application programming interface; AUC: Area under the curve; BERT: Bidirectional encoder
representations from transformers; BLEU: Bilingual evaluation understudy; CM: Cardiomyocytes; EHR: Electronic health record; FLOPs: Floating
point operations per second; iPSC-CM: Induced pluripotent stem cell-derived cardiomyocytes; LIME: Local interpretable model-agnostic explanations;
LLM: Large language model; LMIC: Low- and middle-income countries; NDCG: Normalized discounted cumulative gain; NER: Named entity
recognition; PAGA: Partition-based graph abstraction; PPV: Positive predictive value; Q&A: Question and answer; RMSD: Root mean square deviation;
ROC: Receiver operating characteristic; ROUGE: Recall-oriented understudy for Gisting evaluation; SHAP: Shapley additive explanation;
SNP: Single-nucleotide polymorphism; TM: Template modeling; TPM: Transcripts per million.
Volume 11 Issue 5 (2025) 18 doi: 10.36922/JCTR025230026