Page 65 - JCTR-11-5
P. 65
Journal of Clinical and
Translational Research Metabolism of healthy and leukemic stem cells
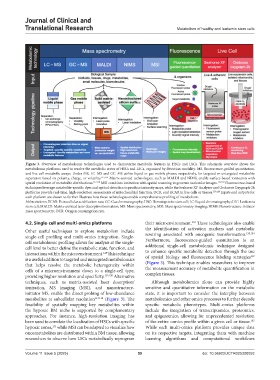
Figure 3. Overview of metabolomic technologies used to characterize metabolic features in HSCs and LSCs. This schematic overview shows the
metabolomic platforms used to resolve the metabolic states of HSCs and LSCs, organized by detection modality: MS, fluorescence-guided quantitation,
and live-cell metabolic assays. Under MS, LC–MS and GC–MS utilize liquid or gas mobile phases, respectively, for targeted or untargeted metabolite
separation based on polarity, charge, or volatility. 115,116 Matrix-assisted technologies, such as MALDI and NIMS, enable surface-based ionization with
spatial resolution of metabolite distributions. 117,118 MSI combines ionization with spatial scanning to generate molecular images. 117,118 Fluorescence-based
techniques leverage metabolite-specific dyes and optical detection to produce intensity maps, while the Seahorse XF Analyzer and Oroboros Oxygraph-2k
platforms provide real-time, high-resolution assessments of mitochondrial function, OCR, and ECAR in live cells or tissues. 119,120 Inputs and outputs for
each platform are shown to further illustrate how these technologies enable comprehensive profiling of metabolism.
Abbreviations: ECAR: Extracellular acidification rate; GC: Gas chromatography; HSC: Hematopoietic stem cell; LC: Liquid chromatography; LSC: Leukemia
stem cell; MALDI: Matrix-assisted laser desorption/ionization; MS: Mass spectrometry; MSI: Mass spectrometry imaging; NIMS: Nanostructure-initiator
mass spectrometry; OCR: Oxygen consumption rate.
132
4.2. Single-cell and multi-omics platforms their microenvironment. These technologies also enable
the identification of activation markers and metabolic
Other useful techniques to explore metabolism include 133,134
single-cell profiling and multi-omics integration. Single- rewiring associated with oncogenic transformation.
cell metabolomic profiling allows for analysis at the single- Furthermore, fluorescence-guided quantitation is an
cell level to better define the metabolic state, function, and additional single-cell metabolomic technique designed
128
interactions within the microenvironment. This technique to enhance specific metabolite detection through the use
of spatial biology and fluorescence labeling strategies
119
is a useful addition to targeted and untargeted metabolomics (Figure 3). This technique enables researchers to improve
that helps resolve the metabolic heterogeneity within the measurement accuracy of metabolite quantification in
cells of a microenvironment down to a single-cell type, complex tissues.
providing higher resolution and specificity. 129,130 Alternative
techniques, such as matrix-assisted laser desorption/ Although metabolomics alone can provide highly
ionization, MS imaging (MSI), and nanostructure- sensitive and quantitative information on the metabolic
initiator MS, enable the direct probing of low-abundance state, it is important to consider the interplay between
metabolites at subcellular resolution 117,118 (Figure 3). The metabolomics and other omics processes to further decode
feasibility of spatially mapping key metabolites within specific metabolic phenotypes. Multi-omics platforms
the hypoxic BM niche is supported by complementary include the integration of transcriptomics, proteomics,
approaches. For instance, high-resolution imaging has and epigenomics, allowing for unprecedented resolution
been used to correlate the localization of HSCs with specific of the entire -omics profile within a given cell or tissue.
135
hypoxic zones, while MSI can be adapted to visualize how While each multi-omics platform provides unique data
131
oncometabolites are distributed within BM tissue, allowing on its respective targets, integrating them with machine
researchers to observe how LSCs metabolically reprogram learning algorithms and computational workflows
Volume 11 Issue 5 (2025) 59 doi: 10.36922/JCTR025320053