Page 101 - MI-2-3
P. 101
Microbes & Immunity SARS-CoV-2 complementary classification
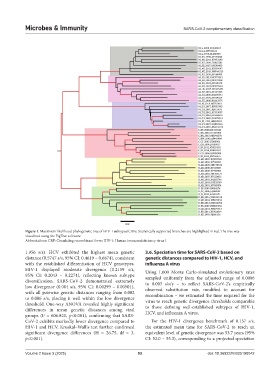
Figure 1. Maximum likelihood phylogenetic tree of HIV-1 subtypes/CRFs. Statistically supported branches are highlighted in red. The tree was
visualized using the FigTree software.
Abbreviations: CRF: Circulating recombinant form; HIV-1: Human immunodeficiency virus 1.
1.956 s/s). HCV exhibited the highest mean genetic 3.6. Speciation time for SARS-CoV-2 based on
distance (0.5747 s/s, 95% CI: 0.4619 – 0.6874), consistent genetic distances compared to HIV-1, HCV, and
with the established differentiation of HCV genotypes. influenza A virus
HIV-1 displayed moderate divergence (0.2159 s/s, Using 1,000 Monte Carlo-simulated evolutionary rates
95% CI: 0.2043 – 0.2274), reflecting known subtype sampled uniformly from the adjusted range of 0.0006
diversification. SARS-CoV-2 demonstrated extremely to 0.003 s/s/y – to reflect SARS-CoV-2’s empirically
low divergence (0.004 s/s, 95% CI: 0.00299 – 0.00501), observed substitution rate, modified to account for
with all pairwise genetic distances ranging from 0.002 recombination – we estimated the time required for the
to 0.006 s/s, placing it well within the low divergence virus to reach genetic divergence thresholds comparable
threshold. One-way ANOVA revealed highly significant
differences in mean genetic distances among viral to those defining well-established subtypes of HIV-1,
HCV, and influenza A virus.
groups (F = 606.821, p<0.001), confirming that SARS-
CoV-2 exhibits markedly lower divergence compared to For the HIV-1 divergence benchmark of 0.157 s/s,
HIV-1 and HCV. Kruskal–Wallis test further confirmed the estimated mean time for SARS-CoV-2 to reach an
significant divergence differences (H = 26.75, df = 3, equivalent level of genetic divergence was 53.7 years (95%
p<0.001). CI: 52.0 – 55.3), corresponding to a projected speciation
Volume 2 Issue 3 (2025) 93 doi: 10.36922/MI025190042