Page 105 - MI-2-3
P. 105
Microbes & Immunity SARS-CoV-2 complementary classification
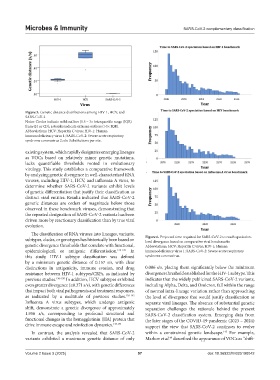
Figure 5. Genetic distance distributions among HIV-1, HCV, and
SARS-CoV-2
Notes: Circles indicate mild outliers (1.5 – 3× interquartile range (IQR)
from Q1 or Q3); asterisks indicate extreme outliers (>3× IQR).
Abbreviations: HCV: Hepatitis C virus; HIV-1: Human
immunodeficiency virus 1; SARS-CoV-2: Severe acute respiratory
syndrome coronavirus 2; s/s: Substitutions per site.
existing system, which rapidly designates emerging lineages
as VOCs based on relatively minor genetic mutations,
lacks quantifiable thresholds rooted in evolutionary
virology. This study establishes a comparative framework
by analyzing genetic divergence in well-characterized RNA
viruses, including HIV-1, HCV, and influenza A virus, to
determine whether SARS-CoV-2 variants exhibit levels
of genetic differentiation that justify their classification as
distinct viral entities. Results indicated that SARS-CoV-2
genetic distances are orders of magnitude below those
observed in these benchmark viruses, demonstrating that
the repeated designation of SARS-CoV-2 variants has been
driven more by reactionary classification than by true viral
evolution.
The classification of RNA viruses into lineages, variants,
subtypes, clades, or genotypes has historically been based on Figure 6. Projected time required for SARS-CoV-2 to reach speciation-
level divergence based on comparative viral benchmarks
genetic divergence thresholds that correlate with functional, Abbreviations: HCV: Hepatitis C virus; HIV-1: Human
epidemiological, or antigenic differentiation. 133-135 In immunodeficiency virus 1; SARS-CoV-2: Severe acute respiratory
this study, HIV-1 subtype classification was defined syndrome coronavirus.
by a minimum genetic distance of 0.157 s/s, with clear
distinctions in antigenicity, immune evasion, and drug 0.006 s/s, placing them significantly below the minimum
resistance between HIV-1 subtypes/CRFs, as indicated by divergence threshold established for the HIV-1 subtype. This
previous studies. 136-138 In addition, HCV subtypes exhibited indicates that the widely publicized SARS-CoV-2 variants,
even greater divergence (≥0.371 s/s), with genetic differences including Alpha, Delta, and Omicron, fall within the range
that impact both viral pathogenesis and treatment responses, of normal intra-lineage variation rather than approaching
as indicated by a multitude of previous studies. 139-142 the level of divergence that would justify classification as
Influenza A virus subtypes, which undergo antigenic separate viral lineages. The absence of substantial genetic
shift, demonstrate a genetic divergence of approximately separation challenges the rationale behind the present
1.956 s/s, corresponding to profound structural and SARS-CoV-2 classification system. Emerging data from
functional changes in the hemagglutinin (HA) protein that the later stages of the COVID-19 pandemic (2023 – 2024)
drive immune escape and reinfection dynamics. 143,144 support the view that SARS-CoV-2 continues to evolve
In contrast, the analysis revealed that SARS-CoV-2 within a constrained genetic landscape. For example,
145
variants exhibited a maximum genetic distance of only Markov et al. described the appearance of VOCs as “shift-
49
Volume 2 Issue 3 (2025) 97 doi: 10.36922/MI025190042