Page 12 - TD-3-1
P. 12
Tumor Discovery Haplotype and LD of BRCA genes in GBM
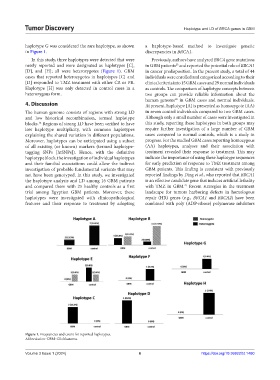
haplotype G was considered the rare haplotype, as shown a haplotype-based method to investigate genetic
in Figure 1. discrepancies in BRCA1.
In this study, three haplotypes were detected that were Previously, authors have analyzed BRCA gene mutations
newly reported and were designated as haplotypes [C], in GBM patients and reported the potential role of BRCA1
25
[D], and [H]; all were heterozygotes (Figure 1). GBM in cancer predisposition. In the present study, a total of 44
cases that reported heterozygotes in haplotypes [C] and individuals were enrolled and categorized according to their
[D] responded to TMZ treatment with either CR or PR. clinical criteria into 15 GBM cases and 29 normal individuals
Haplotype [H] was only detected in control cases in a as controls. The comparison of haplotype concepts between
heterozygous form. two groups can provide reliable information about the
human genome in GBM cases and normal individuals.
26
4. Discussion At present, haplotype [A] is presented as homozygote (AA)
The human genome consists of regions with strong LD in seven control individuals compared to two GBM cases.
and low historical recombination, termed haplotype Although only a small number of cases were investigated in
blocks. Regions of strong LD have been verified to have this study, reporting these haplotypes in both groups may
24
low haplotype multiplicity, with common haplotypes require further investigation of a large number of GBM
explaining the shared variation in different populations. cases compared to normal controls, which is a study in
Moreover, haplotypes can be anticipated using a subset progress. For the studied GBM cases reporting homozygous
of all existing (or known) markers (termed haplotype- (AA) haplotypes, analyses and their association with
tagging SNPs [htSNPs]). Hence, with the definitive treatment revealed their response to treatment. This may
haplotype block, the investigation of individual haplotypes indicate the importance of using these haplotype sequences
and their familial associations could allow the indirect for early prediction of response to TMZ treatment among
investigation of probable fundamental variants that may GBM patients. This finding is consistent with previously
not have been genotyped. In this study, we investigated reported findings by Ding et al., who reported that BRCA1
the haplotype analysis and LD among 16 GBM patients is an effective candidate gene that induces artificial lethality
and compared them with 25 healthy controls as a first with TMZ in GBM. Recent strategies in the treatment
27
trial among Egyptian GBM patients. Moreover, these landscape for tumors harboring defects in homologous
haplotypes were investigated with clinicopathological repair (HR) genes (e.g., BRCA1 and BRCA2) have been
features and their response to treatment by adopting combined with poly (ADP-ribose) polymerase inhibitors
Figure 1. Frequencies and count for reported haplotypes.
Abbreviation: GBM: Glioblastoma.
Volume 3 Issue 1 (2024) 6 https://doi.org/10.36922/td.1480