Page 29 - TD-3-1
P. 29
Tumor Discovery AI uncovers tumor spatial organization
A B C
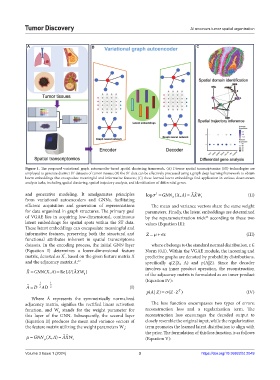
Figure 1. The proposed variational graph autoencoder-based spatial clustering framework. (A) Diverse spatial transcriptomics (ST) technologies are
employed to generate distinct ST datasets of tumor tissues; (B) the ST data can be effectively processed using a graph deep learning framework to obtain
latent embeddings that encapsulate meaningful and informative features; (C) these learned latent embeddings find application in various downstream
analysis tasks, including spatial clustering, spatial trajectory analysis, and identification of differential genes.
and generative modeling. It amalgamates principles log GNNX AXW 1 (II)
2
( ,) A
from variational autoencoders and GNNs, facilitating
efficient acquisition and generation of representations The mean and variance vectors share the same weight
for data organized in graph structures. The primary goal parameters. Finally, the latent embeddings are determined
of VGAE lies in acquiring low-dimensional, continuous by the reparameterization trick according to these two
22
latent embeddings for spatial spots within the ST data. values (Equation III):
These latent embeddings can encapsulate meaningful and
informative features, preserving both the structural and (III)
functional attributes inherent in spatial transcriptome
datasets. In the encoding process, the initial GNN layer where ε belongs to the standard normal distribution, ε ∈
(Equation I) determines a lower-dimensional feature Norm (0,1). Within the VGAE module, the incoming and
matrix, denoted as X ̃, based on the given feature matrix X predictive graphs are denoted by probability distributions,
and the adjacency matrix A: 22 specifically q(Z|X, A) and p(A|Z). Since the decoder
XGNN XA= (, )Re LU AXW( 0 ) involves an inner product operation, the reconstruction
=
of the adjacency matrix is formulated as an inner product
(Equation IV):
1 1
AD AD 2 2 (I)
pA Z(| ) ( Z Z ) T (IV)
Where à represents the symmetrically normalized
adjacency matrix, signifies the rectified linear activation The loss function encompasses two types of errors:
function, and W stands for the weight parameter for reconstruction loss and a regularization term. The
0
this layer of the GNN. Subsequently, the second layer reconstruction loss encourages the decoded output to
(Equation II) produces the mean and variance vectors of closely resemble the original input, while the regularization
the feature matrix utilizing the weight parameters W : term promotes the learned latent distribution to align with
1
the prior. The formulation of this loss function is as follows
GNNX A(, ) AXW 1 (Equation V):
Volume 3 Issue 1 (2024) 3 https://doi.org/10.36922/td.2049