Page 67 - TD-3-4
P. 67
Tumor Discovery RNA-protein complexes deregulated in cancer
Table 3. Gene activation by lncRNAs through euchromatin regulation and histone marks
Gene Interaction Target Histone mark References
GClnc1 WDR5, KAT2A SOD2 promoter H3K4me3, H3K9ac 84
GCAWKR WDR5, KAT2A PTP4A1 H3K4me3, H3K9ac 85
LincRNA‑P21 hnRNP K P21 H3K4me3, H3K9ac 86
TM4SF19‑AS1 WDR5 TM4SF19 H3K4me3 87
TARID GADD45A TCF21 CpG demethylation 88
CCAT1‑L CTCF, hnRNP K MYC Chromatin loop, NUP153 binding to cohesin 89
HOTTIP WDR5, MLL/SET1 HOXA H3K4me3 90
PVT1 CTCF, hnRNP K MYC Chromatin loop, NUP153 binding to cohesin 91
NRIP enhancer Cohesin ER-dependent genes Chromatin loop nuclear receptor-interacting protein 92
HASTER - HNF1A Promoter–enhancer interaction 93,94
lncPRESS1 SIRT6 deacetylase Embryonic state Preserves H3K56ac and H3K9ac 95
NEAT1_2 RBP14 Euchromatin binding Paraspeckle condensates 96
THRIL hnRNPs TNF-α Promoter/enhancer activation 97
HOTAIR LSD1, HBXIP MYC target genes Lysine demethylation 98
NeST WDR5 IFN-gamma H3K4me 99
CircRNA0000285 Unknown FUS involved in back-splicing Cervical cancer, proliferation, metastasis 100
to form circRNAs
circRap1b KAT2A, KAT7 The HOXA5 gene H3K9ac Prevents stroke 101
mark, Fam3a antiapoptotic
Abbreviations: CCAT1-L: Colon cancer-associated transcript 1-long; circRap1b: Circular RNA from Ras-related protein 1b mRNA;
CHD2: Chromodomain helicase DNA-binding protein 2; CpG: Cytosine-phosphate-guanine; Fam3A: Family with sequence similarity 3A, a
cytokine-like protein; FUS: Fused-in sarcoma; HOXA5: Homeobox gene A5; IFN-gamma: Interferon-gamma; MYC: Viral myelocytomatosis homolog;
NEAT1: Nuclear enriched abundant transcript 1; THRIL: TNFα and hnRNPL related immunoregulatory LincRNA; TARID: Tcf21 antisense RNA
inducing demethylation; TNF-α: Tumor necrosis factor-α.
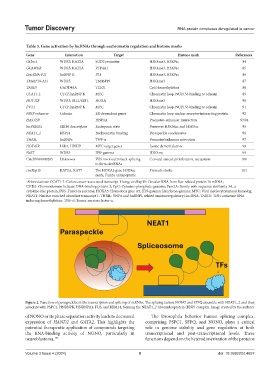
Figure 2. Function of paraspeckles in the transcription and splicing of mRNAs. The splicing factors NONO and SFPQ assemble with NEAT1_2 and then
associate with PSPC1, HNRNPK, HNRNPA1, FUS, and RBM14, forming the NEAT1_2 ribonucleoprotein (RNP) complex. Image created by the authors
of NONO or its phase separation activity leads to decreased The Drosophila behavior human splicing complex,
expression of HAND2 and GATA2. This highlights the comprising PSPC1, SFPQ, and NONO, plays a critical
potential therapeutic application of compounds targeting role in genome stability and gene regulation at both
the RNA-binding activity of NONO, particularly in transcriptional and post-transcriptional levels. These
neuroblastoma. 123 functions depend on the heterodimerization of the proteins
Volume 3 Issue 4 (2024) 8 doi: 10.36922/td.4657