Page 128 - AIH-2-4
P. 128
Artificial Intelligence in Health RefSAM3D for medical image segmentation
the introduction of an efficient pairing attention module. other approaches across a wide range of tasks, achieving
In addition, we compared our approach with 3D UNet- the highest scores in nearly all scenarios, particularly
eXpanded Network (UX-Net), a method designed to excelling in challenging tumor types. In kidney tumor
55
create a simple, efficient, and lightweight network that segmentation, despite challenges such as low contrast
combines the capabilities of hierarchical transformers with with surrounding tissues, blurred boundaries, and high
the advantages of ConvNet modules. We also evaluated morphological heterogeneity, Ref-SAM3D achieved a dice
SAM-B, which is the base model of SAM trained on natural score of 95.53% and an NSD of 99.45%, surpassing other
images and directly applied to medical images without methods. For pancreatic tumors, which constitute less than
adaptation. Finally, our method was benchmarked against 0.5% of CT images and exhibit diverse shapes, Ref-SAM3D
the latest SAM adaptation techniques, including 3DSAM- achieved a dice score of 82.42%, representing a 2.12%
adapter —a promptable 3D medical image segmentation improvement over existing state-of-the-art techniques.
13
model—and MA-SAM, a framework that utilizes In liver tumor segmentation, Ref-SAM3D attained a dice
34
parameter-efficient fine-tuning strategies and 3D adapters. score of 80.10%, effectively handling variations in grayscale
The results presented in Table 2 demonstrate that our and irregular shapes. Despite the extensive distribution and
proposed Ref-SAM3D method consistently outperformed complex anatomical structure of colorectal cancer lesions,
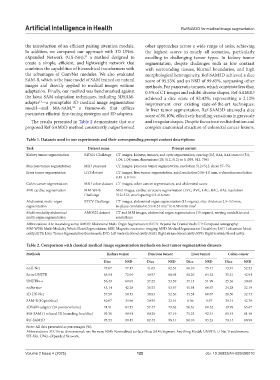
Table 1. Datasets used in our experiments and their corresponding prompt content descriptions
Task Dataset name Prompt content
Kidney tumor segmentation KiTS21 Challenge CT images, kidneys, tumors, and cysts segmentation, spacing (0.5, 0.44, 0.44) mm to (5.0,
1.04, 1.04) mm, dimensions (29, 512, 512) to (1,059, 512, 796)
Pancreas tumor segmentation MSD pancreas CT images, pancreas tumor segmentation, resolution 512×512, slices 37–751
Liver tumor segmentation LiTS dataset CT images, liver tumor segmentation, axial resolution 0.56–1.0 mm, z-direction resolution
0.45–6.0 mm
Colon cancer segmentation MSD colon dataset CT images, colon cancer segmentation, and abdominal scans
MRI cardiac segmentation MM-WHS MRI images, cardiac structure segmentation (LVC, RVC, LAC, RAC, AA), resolution
Challenge 512×512, voxel spacing 0.3–0.6 mm
Abdominal multi-organ BTCV Challenge CT images, abdominal organ segmentation (13 organs), slice thickness 2.5–5.0 mm,
segmentation in-plane resolution 0.54×0.54 mm² to 0.98×0.98 mm²
Multi-modality abdominal AMOS22 dataset CT and MRI images, abdominal organ segmentation (15 organs), varying modalities and
multi-organ segmentation resolutions
Abbreviations: AA: Ascending aorta; AMOS: Abdominal Multi-Organ Segmentation; BTCV: Beyond the Cranial Vault; CT: Computed tomography;
MM-WHS: Multi-Modality Whole Heart Segmentation; MRI: Magnetic resonance imaging; MSD: Medical Segmentation Decathlon; LAC: Left atrium blood
cavity; LiTS: Liver Tumor Segmentation Benchmark; LVC: Left ventricle blood cavity; RAC: Right atrium blood cavity; RVC: Right ventricle blood cavity.
Table 2. Comparison with classical medical image segmentation methods on four tumor segmentation datasets
Methods Kidney tumor Pancreas tumor Liver tumor Colon cancer
Dice NSD Dice NSD Dice NSD Dice NSD
nnU-Net 73.07 77.47 41.65 62.54 60.10 75.41 43.91 52.52
Swin-UNETR 65.54 72.04 40.57 60.05 50.26 64.32 35.21 42.94
UNETR++ 56.49 60.04 37.25 53.59 37.13 51.99 25.36 30.68
nnFormer 45.14 42.28 36.53 53.97 45.54 60.67 24.28 32.19
3D UX-Net 57.59 58.55 34.83 52.56 45.54 60.67 28.50 32.73
SAM-B (10 pts/slice) 40.07 34.96 30.55 32.91 8.56 5.97 39.14 42.70
3DSAM-adapter (10 points/volume) 74.91 84.35 57.47 79.62 56.61 69.52 49.99 65.67
MA-SAM (1 relaxed 3D bounding box/slice) 93.38 98.91 80.30 97.19 75.23 92.31 65.45 81.40
Ref-SAM3D 95.53 99.45 82.42 98.41 80.10 93.23 70.14 88.90
Note: All data presented as percentages (%).
Abbreviations: 3D: Three-dimensional; nn: No new; NSD: Normalized surface Dice; SAM: Segment Anything Model; UNETR: U-Net Transformers;
UX-Net: UNet-eXpanded Network.
Volume 2 Issue 4 (2025) 122 doi: 10.36922/AIH025080010