Page 33 - GPD-2-3
P. 33
Gene & Protein in Disease Signatures construction strategies for TC
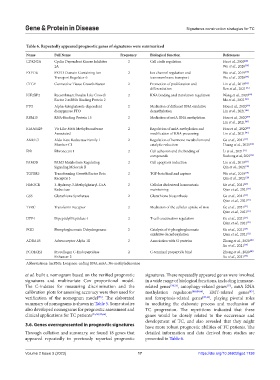
Table 6. Repeatedly appeared prognostic genes of signatures were summarized
Name Full Name Frequency Biological function References
CDKN2A Cyclin Dependent Kinase Inhibitor 2 Cell circle regulation Hu et al., 2020 [22]
2A Wu et al., 2020 [38]
FXYD6 FXYD Domain Containing Ion 2 Ion channel regulation and Wu et al., 2019 [19]
Transport Regulator 6 transmembrane transport Wu et al., 2020 [38]
CTGF Connective Tissue Growth Factor 2 Promotion of proliferation and Lin et al., 2019 [24]
differentiation Ren et al., 2021 [35]
IGF2BP2 Recombinant Insulin Like Growth 2 RNA binding and translation regulation Wang et al., 2020 [66]
Factor 2 mRNA Binding Protein 2 Ma et al., 2021 [30]
FTO Alpha-ketoglutarate-dependent 2 Mediation of different RNA oxidative Hou et al., 2020 [49]
dioxygenase FTO demethylation Lin et al., 2021 [46]
RBM15 RNA-Binding Protein 15 2 Mediation of m6A RNA methylation Hou et al., 2020 [49]
Lin et al., 2021 [46]
KIAA1429 Vir Like M6A Methyltransferase 2 Regulation of m6A methylation and Hou et al., 2020 [49]
Associated modification of RNA processing Lin et al., 2021 [46]
AKR1C1 Aldo-Keto Reductase Family 1 2 Regulation of hormone metabolism and Ge et al., 2021 [27]
Member C1 catalytic reduction Huang et al., 2021 [34]
FN1 Fibronectin 1 2 Cell adhesion and the binding of Li et al., 2021 [27]
compounds Ruchong et al., 2021 [36]
FAM3B FAM3 Metabolism Regulating 2 Cell apoptosis induction Lin et al., 2019 [24]
Signaling Molecule B Qin et al., 2021 [25]
TGFBR3 Transforming Growth Factor Beta 2 TGF-beta bind and capture Wu et al., 2019 [19]
Receptor 3 Qin et al., 2021 [25]
HMGCR 3-Hydroxy-3-Methylglutaryl-CoA 2 Cellular cholesterol homeostasis Ge et al., 2021 [27]
Reductase maintaining Qian et al., 2021 [31]
GSS Glutathione Synthetase 2 Glutathione biosynthesis Ge et al., 2021 [27]
Qian et al., 2021 [31]
TFRC Transferrin Receptor 2 Mediation of the cellular uptake of iron Ge et al., 2021 [27]
Qian et al., 2021 [31]
DPP4 Dipeptidyl Peptidase 4 2 T-cell coactivation regulation Ge et al., 2021 [27]
Qian et al., 2021 [31]
PGD Phosphogluconate Dehydrogenase 2 Catalysis of 6-phosphogluconate Ge et al., 2021 [27]
oxidative decarboxylation Qian et al., 2021 [31]
ADRA1B Adrenoceptor Alpha 1B 2 Association with G proteins Zhong et al., 2020 [40]
Xu et al., 2021 [39]
PCOLCE2 Procollagen C-Endopeptidase 2 C-terminal propeptide bind Zhong et al., 2020 [40]
Enhancer 2 Xu et al., 2021 [39]
Abbreviations: lncRNA: Long non-coding RNA; m6A: N6-methyladenosine
et al. built a nomogram based on the verified prognostic signatures. These repeatedly appeared genes were involved
signatures and multivariate Cox proportional model. in a wide range of biological functions, including immune-
The C-indexes for measuring discrimination and the related genes [24,25] , autophagy-related genes , m6A RNA
[22]
calibration plots for assessing accuracy were then used for methylation regulators [48,49,66] , EMT-related genes ,
[45]
verification of the nomogram model . The elaborated and ferroptosis-related genes [27,31] , playing pivotal roles
[56]
summary of nomograms is shown in Table 5. Some studies in mediating the elaborate process and mechanism of
also developed nomograms for prognostic assessment and TC progression. The repetitions indicated that these
clinical applications for TC patients [25,30,34,66] . genes would be closely related to the occurrence and
development of TC, and also revealed that they might
3.6. Genes overrepresented in prognostic signatures have more robust prognostic abilities of TC patients. The
Through collation and summary, we found 18 genes that detailed information and data derived from studies are
appeared repeatedly in previously reported prognostic presented in Table 6.
Volume 2 Issue 3 (2023) 17 https://doi.org/10.36922/gpd.1138