Page 16 - GPD-4-1
P. 16
Gene & Protein in Disease Gene fusions and chimeric RNAs
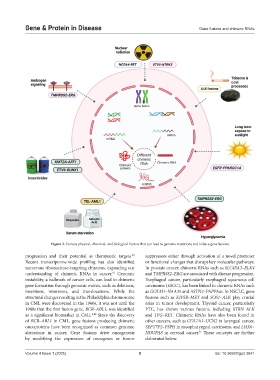
Figure 3. Various physical, chemical, and biological factors that can lead to genome mutations and induce gene fusions
progression and their potential as therapeutic targets. suppressors either through activation of a novel promoter
50
Recent transcriptome-wide profiling has also identified or functional changes that disrupt key molecular pathways.
numerous ribonuclease-targeting chimeras, expanding our In prostate cancer, chimeric RNAs such as SLC45A3–ELK4
understanding of chimeric RNAs in cancer. Genomic and TMPRSS2–ERG are associated with disease progression.
51
instability, a hallmark of cancer cells, can lead to chimeric Esophageal cancer, particularly esophageal squamous cell
gene formation through genomic events, such as deletions, carcinoma (ESCC), has been linked to chimeric RNAs such
insertions, inversions, and translocations. While the as GOLM1–NAA35 and ASTN2–PAPPAas. In NSCLC, gene
structural changes resulting in the Philadelphia chromosome fusions such as KIF5B–MET and SOS1–ALK play crucial
in CML were discovered in the 1960s, it was not until the roles in tumor development. Thyroid cancer, particularly
1980s that the first fusion gene, BCR–ABL1, was identified PTC, has shown various fusions, including STRN–ALK
as a significant biomarker in CML. Since the discovery and TFG–RET. Chimeric RNAs have also been found in
6,48
of BCR–ABL1 in CML, gene fusions producing chimeric other cancers, such as COL7A1–UCN2 in laryngeal cancer,
oncoproteins have been recognized as common genomic SEPT7P2–PSPH in nasopharyngeal carcinoma, and LHX6–
alterations in cancer. Gene fusions drive oncogenesis NDUFA8 in cervical cancer. These concepts are further
51
by modifying the expression of oncogenes or tumor elaborated below.
Volume 4 Issue 1 (2025) 5 doi: 10.36922/gpd.3641