Page 21 - GPD-4-1
P. 21
Gene & Protein in Disease Gene fusions and chimeric RNAs
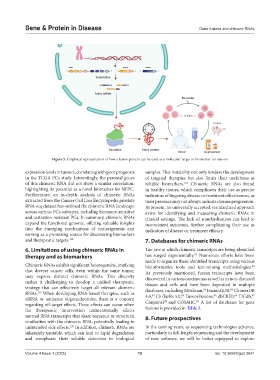
Figure 5. Graphical representation of how a fusion protein can be used as a molecular target or biomarker for tumors
expression levels in tumors, correlating with poor prognosis samples. This instability not only hinders the development
in the TCGA PCa study. Interestingly, the parental genes of targeted therapies but also limits their usefulness as
of this chimeric RNA did not show a similar correlation, reliable biomarkers. Chimeric RNAs are also found
121
highlighting its potential as a novel biomarker for NEPC. in healthy tissues, which complicates their use as precise
Furthermore, an in-depth analysis of chimeric RNAs indicators of lingering disease or treatment effectiveness, as
extracted from the Cancer Cell Line Encyclopedia prostate their presence may not always indicate disease progression.
RNA-seq dataset has outlined the chimeric RNA landscape At present, no universally accepted, standardized approach
across various PCa subtypes, including hormone-sensitive exists for identifying and measuring chimeric RNAs in
and castration-resistant PCa. In summary, chimeric RNAs clinical settings. The lack of standardization can lead to
expand the functional genome, offering valuable insights inconsistent outcomes, further complicating their use as
into the emerging mechanisms of tumorigenesis and indicators of disease or treatment efficacy.
serving as a promising source for discovering biomarkers
and therapeutic targets. 118 7. Databases for chimeric RNAs
6. Limitations of using chimeric RNAs in The rate at which chimeric transcripts are being identified
79
therapy and as biomarkers has surged exponentially. Numerous efforts have been
made to organize these identified transcripts using various
Chimeric RNAs exhibit significant heterogeneity, implying bioinformatics tools and text-mining methodologies.
80
that diverse cancer cells, even within the same tumor, As previously mentioned, fusion transcripts have been
may express distinct chimeric RNAs. This diversity discovered in various carcinomas as well as in non-diseased
makes it challenging to develop a unified therapeutic tissues and cells and have been deposited in multiple
strategy that can effectively target all relevant chimeric databases, including Mitelman, FusionGDB, ChimerDB
82
81
RNAs. When developing RNA-based therapies, such as 4.0, ChiTarRs 5.0, TumorFusions, dbCRID, TICdb,
119
87
83
85
86
84
siRNA or antisense oligonucleotides, there is a concern 88 89
regarding off-target effects. These effects can occur when ConjoinG and COSMIC. A list of databases for gene
the therapeutic intervention unintentionally affects fusions is provided in Table 3.
normal RNA transcripts that share sequence or structural 8. Future prospectives
similarities with the chimeric RNA, potentially leading to
unintended side effects. In addition, chimeric RNAs are In the coming years, as sequencing technologies advance,
120
inherently unstable, which can lead to rapid degradation particularly in full-length sequencing and the development
and complicate their reliable detection in biological of new software, we will be better equipped to explore
Volume 4 Issue 1 (2025) 10 doi: 10.36922/gpd.3641