Page 120 - ITPS-7-2
P. 120
INNOSC Theranostics and
Pharmacological Sciences PI3K-α inhibitors for cancer immunotherapy
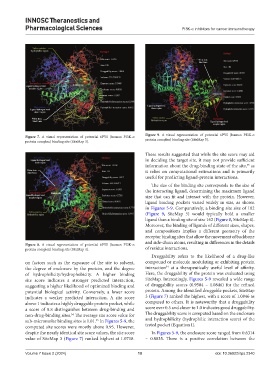
Figure 7. A visual representation of potential 6PYS (human PI3K-α Figure 9. A visual representation of potential 6PYS (human PI3K-α
protein complex) binding site (SiteMap 3). protein complex) binding site (SiteMap 5).
These results suggested that while the site score may aid
in deciding the target site, it may not provide sufficient
55
information about the drug-binding state of the site, as
it relies on computational estimations and is primarily
useful for predicting ligand-protein interactions.
The size of the binding site corresponds to the size of
the interacting ligand, determining the maximum ligand
size that can fit and interact with the protein. However,
ligand binding pockets varied widely in size, as shown
in Figures 5-9. Comparatively, a binding site size of 102
(Figure 9, SiteMap 5) would typically hold a smaller
ligand than a binding site of size 162 (Figure 8, SiteMap 4).
Moreover, the binding of ligands of different sizes, shapes,
and compositions implies a different geometry of the
receptor binding sites that allow the movement of backbone
and side-chain atoms, resulting in differences in the details
Figure 8. A visual representation of potential 6PYS (human PI3K-α
protein complex) binding site (SiteMap 4). of residue interactions.
Druggability refers to the likelihood of a drug-like
on factors such as the exposure of the site to solvent, compound or molecule modulating or exhibiting protein
54
the degree of enclosure by the protein, and the degree interaction at a therapeutically useful level of affinity.
of hydrophilicity/hydrophobicity. A higher binding Here, the druggability of the protein was evaluated using
site score indicates a stronger predicted interaction, SiteMap. Interestingly, Figures 5-9 revealed a wide range
suggesting a higher likelihood of optimized binding and of druggability scores (0.9504 – 1.0846) for the refined
potential biological activity. Conversely, a lower score protein. Among the identified druggable pockets, SiteMap
indicates a weaker predicted interaction. A site score 3 (Figure 7) ranked the highest, with a score of 1.0846 as
above 1 indicates a highly druggable protein pocket, while compared to others. It is noteworthy that a druggability
a score of 0.8 distinguishes between drug-binding and score over 0.5 and closer to 1.0 indicates good druggability.
non-drug-binding sites. The average site score value for The druggability score is computed based on the enclosure
54
sub-micromolar binding sites is 1.01. In Figures 5-9, the and hydrophilicity (hydrophilic interaction score) of the
55
computed site scores were mostly above 0.95. However, tested pocket (Equation I).
despite the nearly identical site score values, the site score In Figures 5-9, the enclosure score ranged from 0.6314
value of SiteMap 3 (Figure 7) ranked highest at 1.0718. – 0.8035. There is a positive correlation between the
Volume 7 Issue 2 (2024) 10 doi: 10.36922/itps.2340