Page 122 - ITPS-7-2
P. 122
INNOSC Theranostics and
Pharmacological Sciences PI3K-α inhibitors for cancer immunotherapy
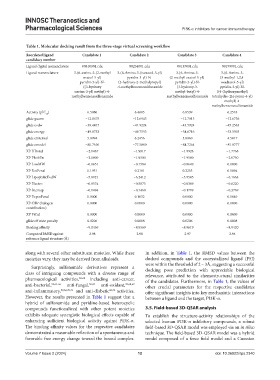
Table 1. Molecular docking result from the three‑stage virtual screening workflow
Reordered ligand Candidate 1 Candidate 2 Candidate 3 Candidate 4
candidacy number
Ligand digital nomenclature 00310001.cdx 00254001.cdx 00137001.cdx 00199001.cdx
Ligand nomenclature 3-(6-amino-5-(2-methyl 3-(6-Amino-5-(isoxazol-5-yl) 3-[6-Amino-5- 3-(6-Amino-5-
oxazol-5-yl) pyridin-3-yl)-N- (2-methyl-oxazol-5-yl)- (3-methyl-1,2,4-
pyridin-3-yl)-N- (2-hydroxy-2-methylpropyl) pyridin-3-yl]-N- oxadiazol-5-yl)
((3-hydroxy -4-methylbenzenesulfonamide (3-hydroxy-3- pyridin-3-yl)-N-
oxetan-3-yl) methyl)-4- methyl-butyl)-4- ((4-(hydroxymethyl)
methylbenzenesulfonamide methylbenzenesulfonamide tetrahydro-2H-pyran-4-yl)
methyl)-4-
methylbenzenesulfonamide
Activity (pIC ) 6.5086 6.4685 6.8539 6.2518
50
glide gscore −12.8675 −12.6943 −12.7015 −12.6736
glide evdw −38.4417 −41.9224 −43.5924 −45.2563
glide energy −49.8733 −49.7553 −54.6716 −53.5503
glide einternal 5.0994 6.2456 5.8060 4.5817
glide emodel −80.7610 −77.5800 −84.7264 −81.8777
XP HBond −2.1467 −1.9817 −1.9926 −1.7768
XP PhobEn −2.0000 −1.9500 −1.9500 −2.0750
XP LowMW −0.0651 −0.1584 −0.0649 0.0000
XP RotPenal 0.1931 0.2181 0.2253 0.1894
XP LipophilicEvdW −5.8921 −6.2412 −5.9585 −6.1663
XP Electro −0.8574 −0.5875 −0.8309 −0.6220
XP Sitemap −0.1494 −0.1468 −0.1799 −0.3769
XP ExposPenal 0.0000 0.1032 0.0000 0.1040
XP ClBr (halogen 0.0000 0.0000 0.0000 0.0000
contribution)
XP PiCat 0.0000 0.0000 0.0000 0.0000
glide eff state penalty 0.0206 0.0008 0.0206 0.0008
Binding affinity −9.0456 −8.9569 −8.9619 −8.9420
Computed RMSD against 2.98 2.98 2.97 2.94
reference ligand structure (Å)
along with several other substituent moieties. While these In addition, in Table 1, the RMSD values between the
moieties vary, they may be derived from alkaloids. docked compounds and the cocrystallized ligand (P5J)
were within the threshold of 2 – 3Å, suggesting a successful
Surprisingly, sulfonamide derivatives represent a docking pose prediction with appreciable biological
class of intriguing compounds with a diverse range of relevance, attributed to the chemostructural similarities
pharmacological activities, 58-60 including anti-cancer, of the candidates. Furthermore, in Table 1, the values of
anti-bacterial, 59,61-66 anti-fungal, 59,67 anti-oxidant, 59,68,69 other crucial parameters for the respective candidates
anti-inflammatory, 59,62,70,71 and anti-diabetic 60,72 activities. offer significant insights into key mechanistic interactions
However, the results presented in Table 1 suggest that a between a ligand and the target, PI3K-α.
hybrid of sulfonamide and pyridine-based heterocyclic
compounds functionalized with other potent moieties 3.5. Field-based 3D-QSAR analysis
exhibits adequate synergistic biological effects capable of To establish the structure-activity relationships of the
enhancing sufficient biological activity against PI3K-α. selected human PI3K-α inhibitory compounds, a robust
The binding affinity values for the respective candidates field-based 3D-QSAR model was employed via an in silico
demonstrated a reasonable reflection of a spontaneous and technique. The field-based 3D-QSAR model was a hybrid
favorable free energy change toward the bound complex. model composed of a force field model and a Gaussian
Volume 7 Issue 2 (2024) 12 doi: 10.36922/itps.2340